Figure 5.
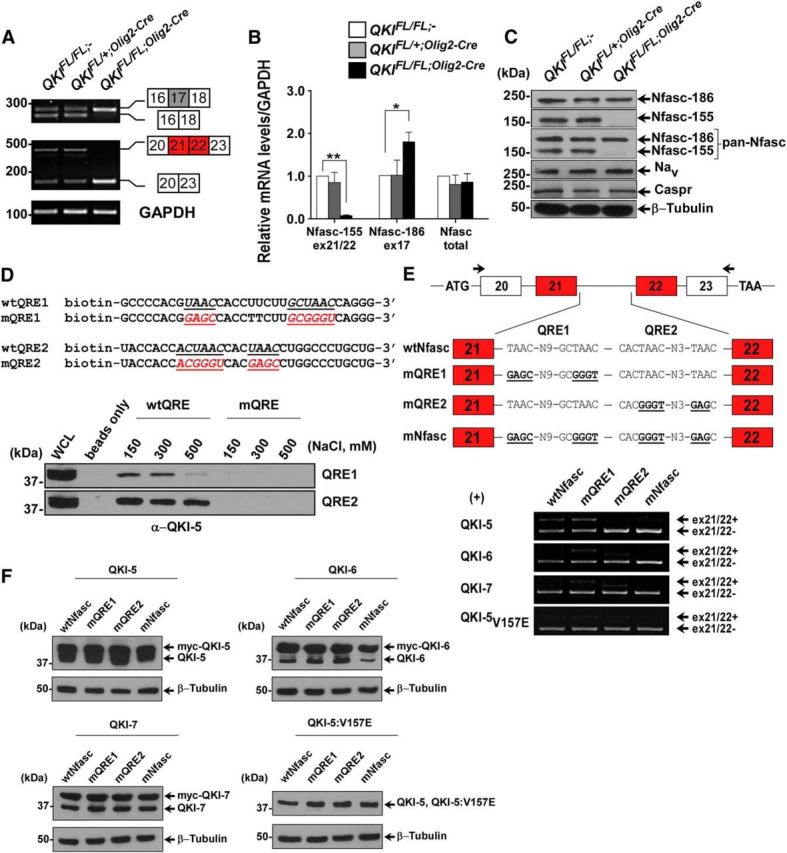
QKI regulates alternative splicing of Nfasc pre-mRNA. A, RT-PCR analysis from P14 mice brains with the indicated primers flanking exon 17 (inclusion denotes Nfasc186) and exons 21/22 (inclusion denotes Nfasc155). B, RT-qPCR using primers spanning the exon–exon junction for exon 17 of Nfasc186, exons 21/22 of Nfasc155, and primers that detect the total mRNA of Nfasc. The results are represented in terms of fold change after normalizing the mRNA levels to GAPDH mRNA from n = 3. Each value represents the mean ± SEM. *p < 0.05; **p < 0.01. C, Western blot analysis of total brain lysates immunoblotted with anti-Nfasc155, anti-Nfasc186, anti-pan-Nfasc, anti-Nav, anti-Caspr, and anti-β-tubulin. Molecular mass markers are shown on the left in kilodaltons. D, Immunoblot analysis of RNA pull-down assay from brains of wild-type P14 mice using biotinylated RNA harboring either wild-type or mutated QREs (mutated sequences indicated) using QKI-5 antibody. Increasing salt concentrations were used to compete with the binding. E, Schematics of the genomic DNA region used to generate the Nfasc155 minigene; the location of the primer set used for RT-PCR are marked by arrows. Wild-type sequences as well as sequences harboring mutant QRE1, QRE2, or both are shown. HEK293T cells were transfected with the Nfasc minigene plasmid and either expression vectors for QKI-5, QKI-6, QKI-7, or QKI-5:V157E. Total RNA was isolated 48 h later and analyzed by RT-PCR. F, Protein expression of transfected plasmids in HEK293T cells is determined by immunoblotting using the indicated antibodies. Myc-tagged QKI proteins migrate higher than endogenous QKI isoforms.
