Figure 4.
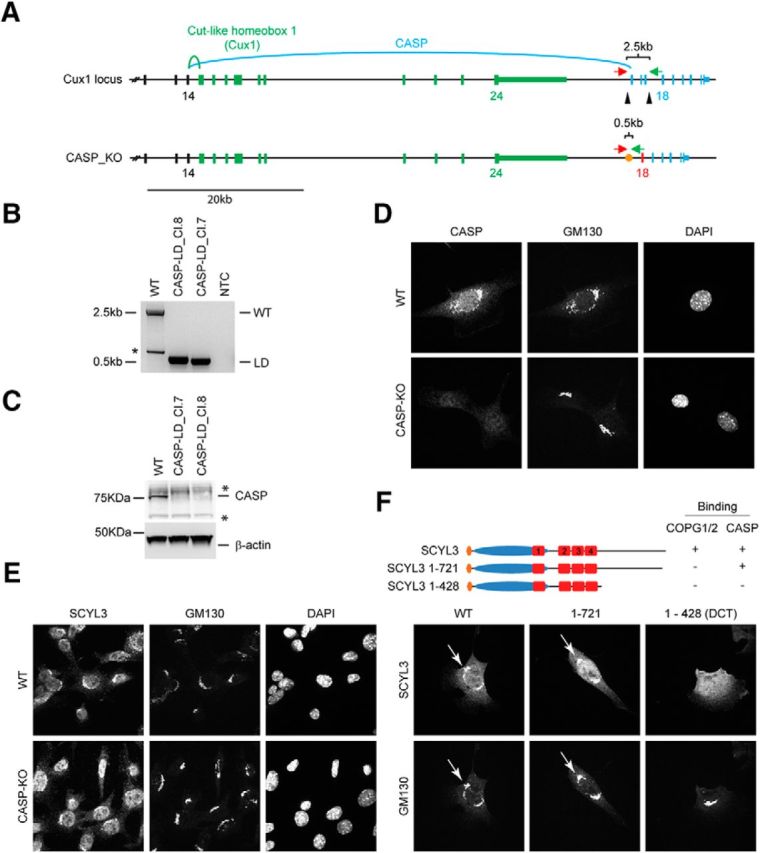
COPI and CASP interactions contribute to localization of SCYL3 to the Golgi apparatus. A, Generation of CASP-deficient MEFs using CRISPR–Cas9 technology. Structure of the Cux1 gene encoding CDP and CASP. Exons common to CDP and CASP are illustrated in black (only exons 12–14 illustrated). Exons encoding CDP are illustrated in green, and those encoding CASP are illustrated in blue. To generate CASP-deficient cells, 2 sgRNAs (black arrowheads) were designed to delete exons 15–17. Splicing of exon 14 into exon 18 causes a frame shift and premature stop codon. Red arrow, Cux1-F51 primer; green arrow, Cux-R32 primer. B, PCR genotyping of the WT and 2 independently derived CASP-deficient MEF lines (Cl.7 and Cl.8). Bands of 2496 and ∼495 bp correspond to WT and null CASP-null alleles, respectively. A smaller band (*) from internal priming sites was also obtained by using these primers. NTC, No template control. Sanger sequencing confirmed proper recombination of the allele in CASP-deficient MEFs. C, Western blot analysis of CASP and β-actin (as loading control) in WT and CASP-deficient MEFs. *Indicates nonspecific bands. D, Immunofluorescence staining of CASP and GM130 in WT and CASP-deficient MEFs. Exponentially growing WT and CASP-deficient MEFs were fixed and stained for CASP and GM130 and imaged by confocal microscopy. E, SCYL3 localization in WT and CASP-deficient MEFs. Exponentially growing WT and CASP-deficient MEFs were fixed and stained for SCYL3 and GM130 and imaged by confocal microscopy. F, Subcellular localization of full-length and key truncated version of SCYL3 in Scyl3-deficient MEFs. Exponentially growing Scyl3−/− MEFs were transfected with full-length or the indicated truncated versions of SCYL3, fixed and stained for SCYL3 and GM130, and imaged by confocal microscopy.
