Figure 6.
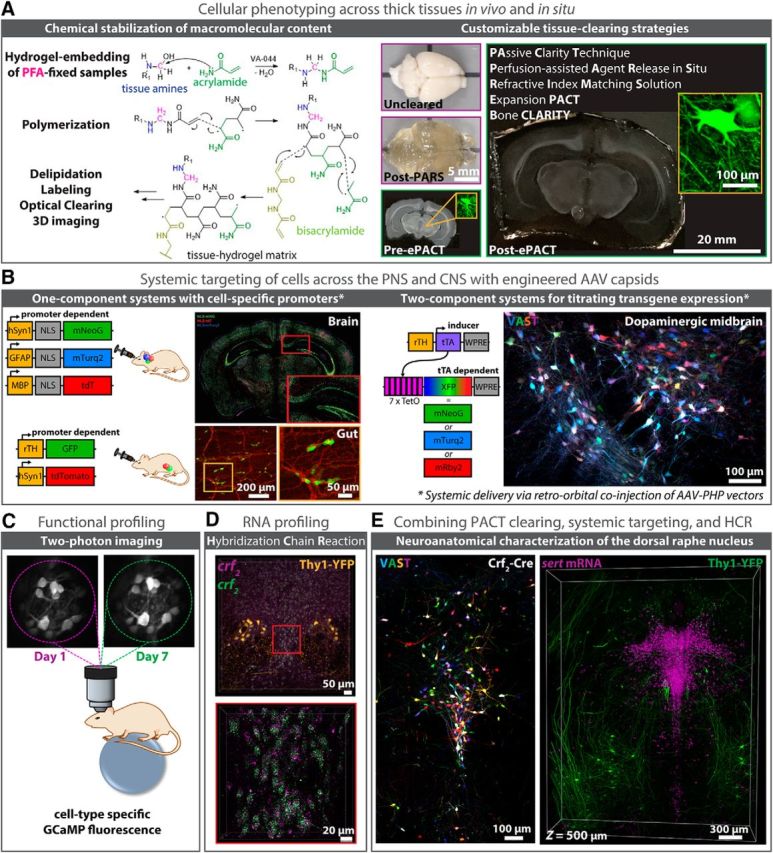
Advancing modern neuroscience endeavors is the development of techniques for modifying chemically-distinct and/or genetically-specified cells and circuits for high-resolution imaging at depth. Among these techniques are as follows: (A) HTC-based biomolecule stabilization and tissue-clearing (images adapted from Yang et al., 2014; Treweek et al., 2015; Treweek and Gradinaru, 2016; Greenbaum et al., 2017a; Gradinaru et al., 2018), (B) viral vector engineering [left, Cre recombinase-based AAV targeted evolution (CREATE); right, vector-assisted spectral tracing (VAST); Deverman et al., 2016; Chan et al., 2017; Bedbrook et al., 2018; Challis et al., 2018], and (C, E) methodologies for cell-profiling in vivo and in situ. Recent advances in modern microscopy [e.g., LSFM and ultramicroscopy (Dodt et al., 2007); CLARITY-optimized light-sheet microscopy (Tomer et al., 2014); two-photon microscopy and microendoscopy (Jung et al., 2004; Barretto et al., 2011; Marshall et al., 2016)], allow researchers to visualize deep tissue structures in real-time (C), and to image large tissue volumes rapidly and with subcellular resolution [D, E; HCR (Choi et al., 2014; Shah et al., 2016; Greenbaum et al., 2017b)]. Complementing these technologies are improved strategies for targeting individual cell populations and neuronal circuits, such as through the use of genetically engineered animal models (e.g., rodent fluorescent reporter and Cre driver lines; D, E) or through the systemic delivery (e.g., via retro-orbital (RO) injection) of viral vectors with unique cell tropism (Deverman et al., 2016; Chan et al., 2017); B, Top left, For CNS targeting, AAV-PHP.eB was used to package single-stranded (ss) rAAV genomes expressing nuclear localized (NLS) fluorescent reporters (XFP) from cell type-specific promoters for neurons (hSyn1, green), oligodendrocytes (MBP, red), or astrocytes (GFAP, blue), resulting in brain-wide gene expression upon RO delivery; bottom left, for PNS targeting, AAV-PHP.S was used to package ss-rAAV genomes expressing XFPs from either neuron-specific (hSyn1) or tyrosine hydroxylase (rTH)-specific promoters, with RO injection of ssAAVPHP.S:rTH-GFP and ssAAV-PHP.S:hSyn1-tdTomato-f (farnesylated) resulting in gene expression throughout TH+-containing cell bodies (green) and nerve bundles (red) of the myenteric and submucosal plexus of the duodenum; images adapted from Challis et al., 2018) and/or multicolor labeling capabilities (Chan et al., 2017; e.g., VAST; B, right; E, left), the latter of which promotes cell-sorting and long-range projection mapping endeavors (Bedbrook et al., 2018). D, E, Likewise, methodologies for labeling RNA transcripts and protein epitopes have been adapted for use alongside tissue-clearing protocols (E; Shah et al., 2016; Greenbaum et al., 2017b). Here, HCR is uniquely suited to imaging RNA point-labels at depth with up to single-molecule precision (D), and to resisting label loss during immunohistochemistry (E) through its use of fluorescence signal amplification and direct in situ hybridization-based readout (Choi et al., 2014).
