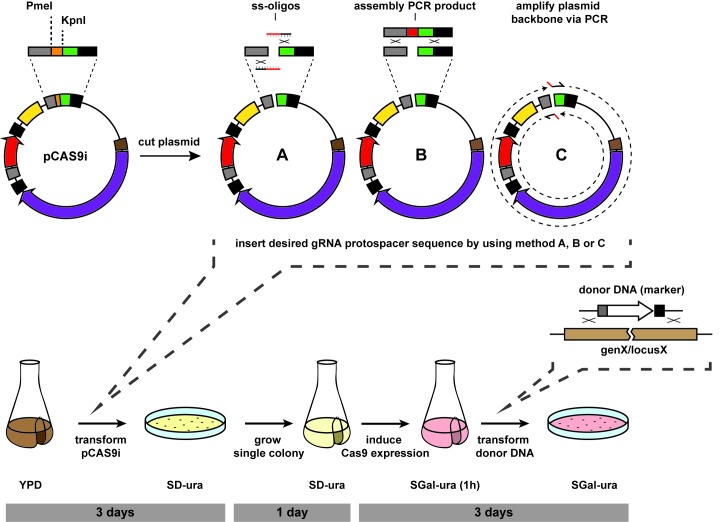
Figure 2. General experimental procedure of pCAS9i-supported CRISPR/Cas9 genome editing in S. cerevisiae.
The stuffer sequence of plasmid pCAS9i is removed by double-cleaving the vector with KpnI and PmeI (top left). A desired protospacer sequence can be introduced into the linearized vector pCAS9i by method A (one-step assembly of overlapping single-stranded oligonucleotides), method B (introduction of a gRNA expression cassette previously generated with assembly PCR) or method C (PCR amplification of pCAS9i backbone with overhang primers that add the protospacer sequence to both ends of the PCR product) (top row). The target strain is (co-)transformed with all plasmid components so that plasmid assembly and recircularization occurs in vivo by recombination-based cloning. Positive transformants are selected on SD-ura agar plates (bottom left). Proper introduction of the desired gRNA protospacer sequence into plasmid pCAS9i can be checked optionally by colony PCR. A single transformant harboring the protospacer containing plasmid (pCAS9i-antiX) is grown overnight in SD-ura liquid medium and used for a second transformation with the respective donor DNA to be genomically integrated. Prior to this second transformation pCAS9-antiX harboring cells are shifted to SGal-ura medium for 1 h in order to induce Cas9 expression from the GAL1 promoter and preloading cells with gRNA and the Cas9 endonuclease. Transformed cells can be selected on appropriate SGal agar medium. If a donor DNA contains a selection marker, cells may be spread on SGal-ura medium that is lacking for a second nutrient (bottom row).