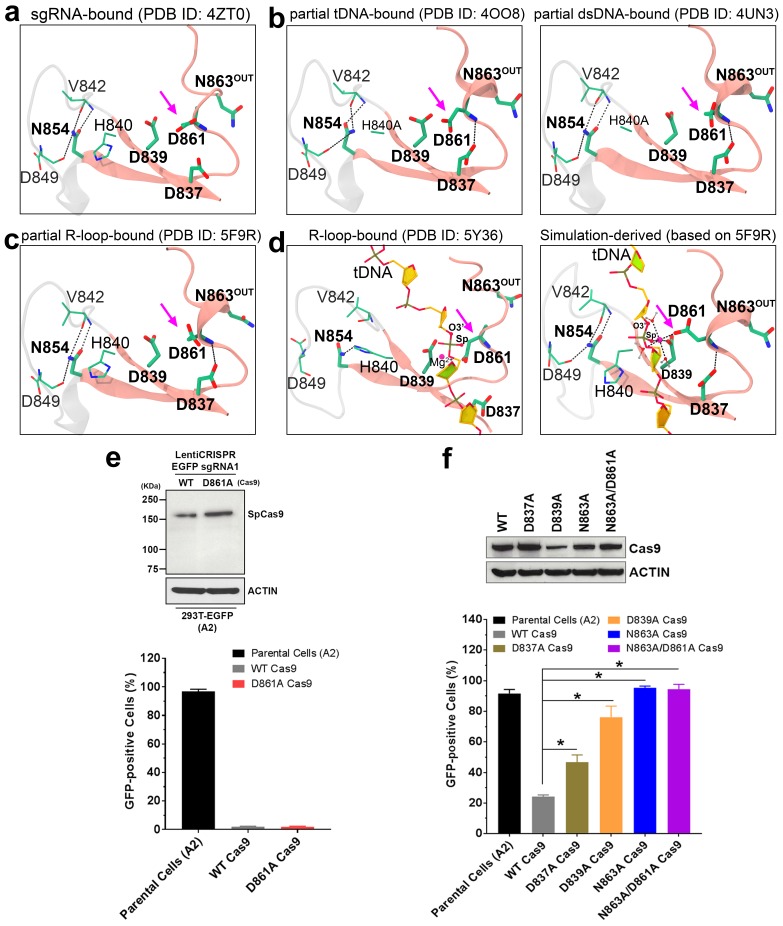
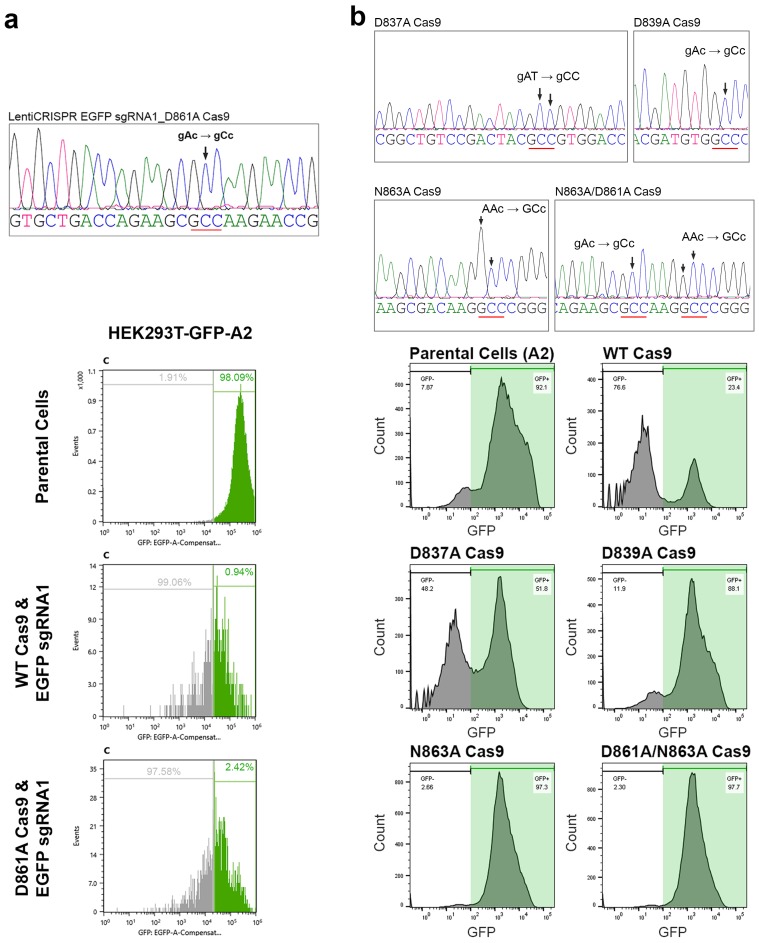
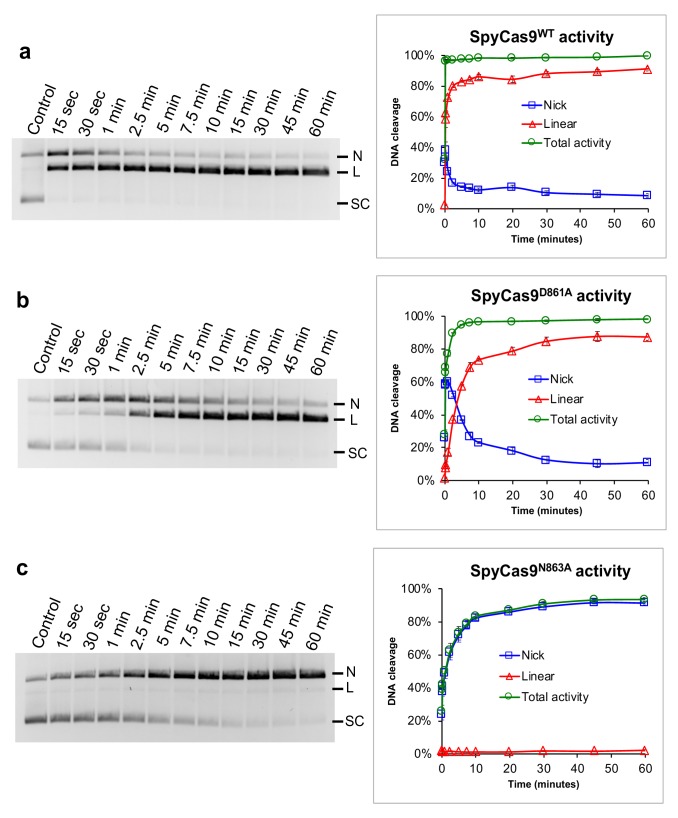
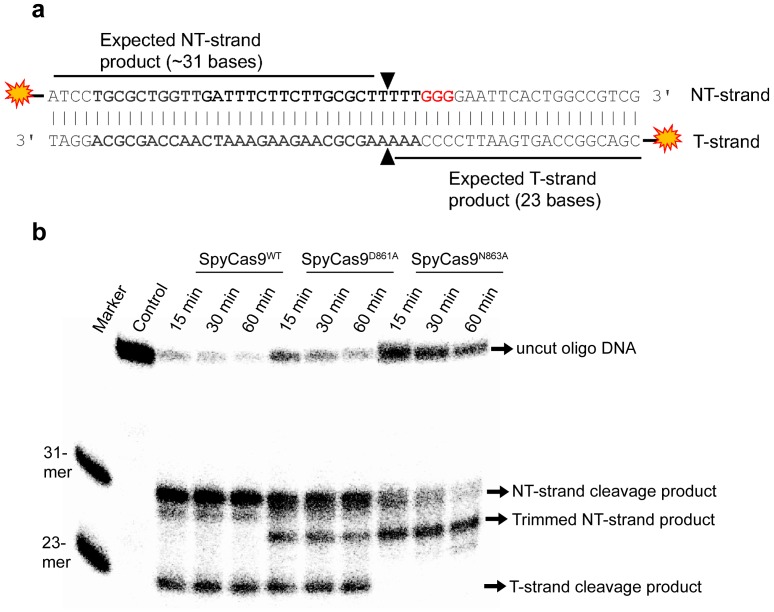
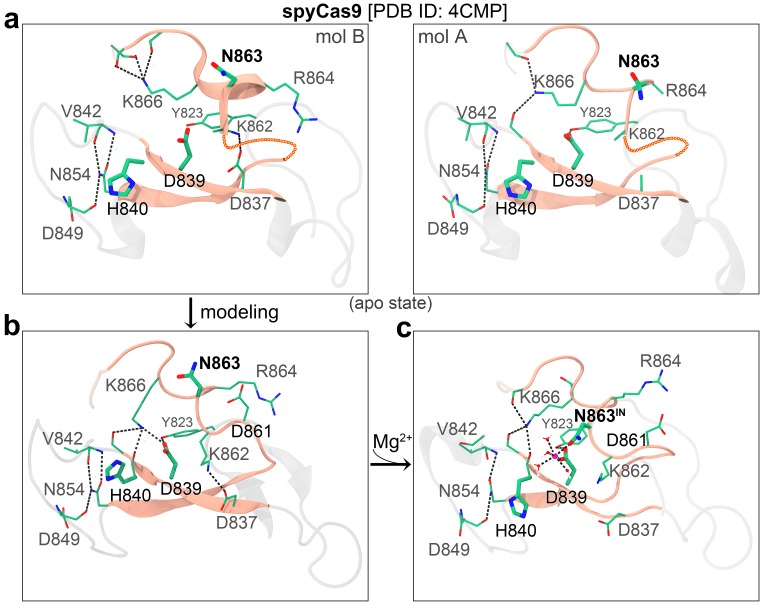
Figure 1. Architecture of the HNH domain ββα-Me fold in different binding forms of Cas9 (a–d) and site-directed mutagenesis experiments identifying potential catalytic residues (e–f).
(a–d) ββα-Me fold in the sgRNA-bound state of Cas9 (a), in the intermediate state (b), in the pre-catalytic state (c), and in the pseudoactive state (d). The ββα-Me fold is represented as pink ribbons, and the residues are shown in stick models and colored by atom type (C, dark green; N, blue; O, red). If present, the bound Mg2+ ion is depicted as a magenta sphere, and only the tDNA phosphate-sugar backbone is displayed for clarity. The location of the Cas9 D861 is highlighted by an arrow, and the dashed lines denote hydrogen bonds or coordinative bonds. (e) The expression and DNA-editing activity of the wild-type and D861A variants of Cas9 paired with an sgRNA sequence that targets the egfp gene in HEK293T-EGFP cells. (f) The expression and DNA-editing activity of the wild-type and indicated variants of Cas9 paired with an sgRNA sequence that targets the egfp gene in HEK293T-EGFP cells. The retention of EGFP expression reflected the loss of activity of Cas9 protein in the cells.