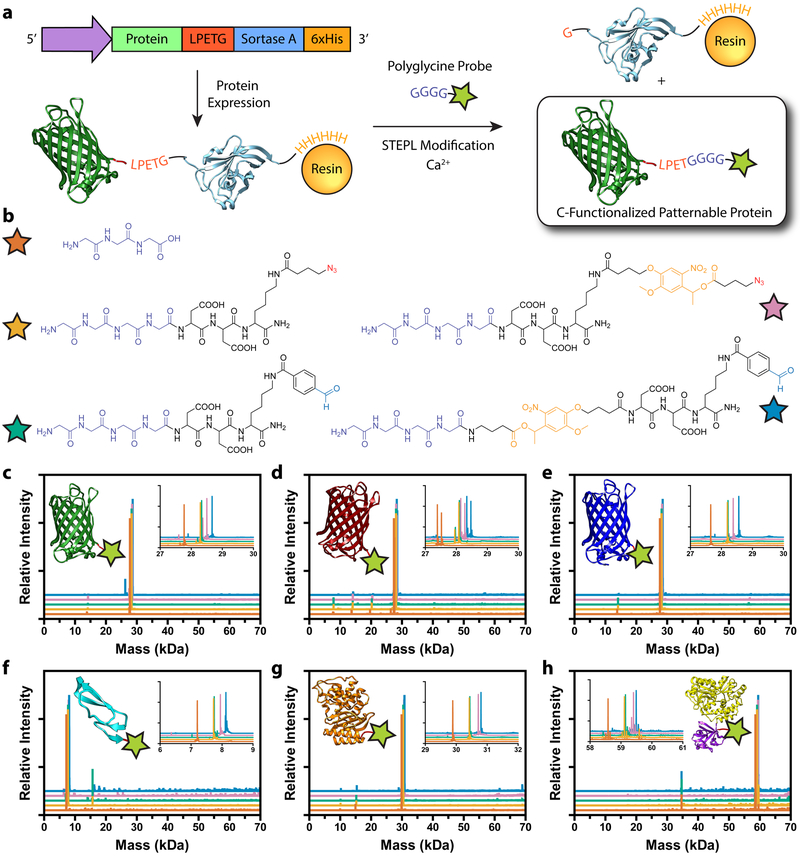
Figure 1 |. Generation of sortagged protein library for biomaterial modification.
a. Sortase-Tag Expressed Protein Ligation (STEPL) enables one-step protein biofunctionalization and purification of C-modified proteins for biomaterial decoration. Proteins appended with a genetically encoded sorting signal are expressed as a fusion with the sortase enzyme and a 6xHis tag. Following chromatographic isolation on Ni-NTA resin, intramolecular sortagging is promoted by the addition of calcium and a polyglycine probe, catalyzing peptide ligation to the protein of interest and simultaneous displacement from the 6xHis-functionalized sortase A. Final protein functionality is defined by polyglycine compound identity. b. Five distinct polyglycine probes each with different reactive functional groups enabling biomaterial decoration were synthesized and exploited for STEPL: triglycine, H-GGGGDDK(N3)-NH2, H-GGGGDDK(CHO)-NH2, H-GGGGDDK(oNB-N3)-NH2, H-GGGG-oNB-DDK(CHO)-NH2 (denoted respectively with orange, tan, teal, pink, and blue stars). c-h. Whole-protein mass spectrometry of sortagged proteins (c, EGFP; d, mCherry; e, mCerulean; f, EGF; g, bla; h, FGF) indicates high sample purity and quantitative functionalization for all polyglycine probes. Curve color denotes probe identity following the same scheme as b. Double peaks in d indicate incomplete N-terminal methionine excision common to mCherry expression. Spectra in c-h correspond to observed masses from a single purification of each species.