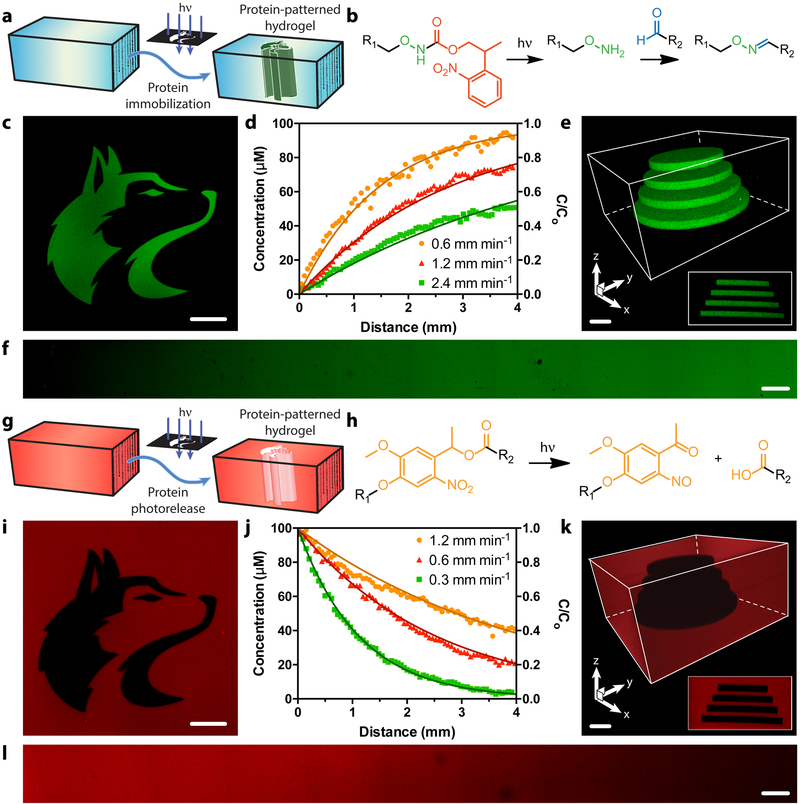
Figure 3 |. Photopatterned alteration of hydrogel biomaterials with sortagged proteins.
a-f. Photopatterned immobilization of sortagged proteins within gels. a. Proteins modified site-specifically with aromatic aldehydes are immobilized within SPAAC-based gels through photomediated oxime ligation. b. NPPOC-caged alkoxyamines distributed uniformly throughout gels (R1) undergo irreversible β-elimination upon mild UV light exposure (λ = 365 nm or 740 nm). The deprotected alkoxyamines react with aldehyde-tagged proteins (R2) to form stable oxime linkages. c. Mask-based photolithography (λ = 365 nm) was used to immobilize discrete patterns of EGFP-CHO throughout the gel thickness. d,f. By exposing gel surfaces to linear gradients of light exposure (created by covering samples with an opaque photomask moving at rates of 0.6, 1.2, 2.4 mm min−1, 10 mW cm−2, λ = 365 nm), exponential protein gradients were generated in a dose-dependent manner. C0 represents the highest possible protein concentration that can be immobilized (determined based on the 100 μM caged alkoxyamine included during gel formulation). e. Full 3D control over protein tethering within gels is achieved through multiphoton laser-scanning lithographic patterning (λ = 740 nm). g-l. Photopatterned release of sortagged proteins from gels. g. Site-specifically modified proteins are released from SPAAC-based gels through ortho-nitrobenzyl ester (oNB) photocleavage. h. oNB moieties linking the sortagged proteins (R1) and the hydrogel (R2) undergo rapid photoscission upon mild UV light exposure (λ = 365 nm or 740 nm). i. Mask-based photolithography (λ = 365 nm) was used to dictate discrete patterns of protein release from gels uniformly functionalized with mCherry-oNB-N3. j,l. By exposing gel surfaces to linear gradients of light exposure (created by covering samples with an opaque photomask moving at rates of 0.3, 0.6, 1.2 mm min−1, 10 mW cm−2, λ = 365 nm), exponential protein gradients were generated in a dose-dependent manner. C0 corresponds to the initial mCherry concentration included during gel formulation (100 μM). k. Full 3D control over protein release within gels is achieved through multiphoton laser-scanning lithographic patterning (λ = 740 nm). Immobilized protein concentrations in d and j are determined through fluorescence correlation. Solid curves in d and j are predicted by known NPPOC/oNB photocleavage kinetics. Images in c, e, f, i, k, and l correspond to representative individual gels imaged by fluorescence confocal microscopy. Data in d and j was derived from experiments involving single gels for each gradient light condition. Inset in e corresponds to a maximum-intensity y-projection, while that in k corresponds to a minimum-intensity y-projection. Scale bars = 100 μm.