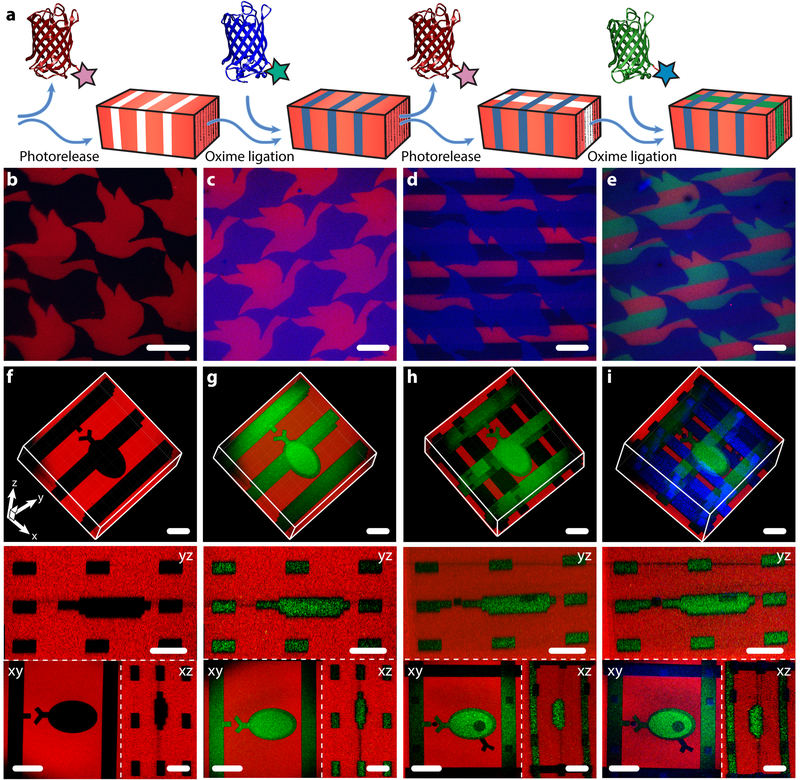
Figure 4 |. 4D photoevolution of hydrogel biomaterials patterned with multiple sortagged proteins.
a. Protein photorelease can be performed in concert with photomediated ligation of an aldehyde-tagged protein to create complex interconnected biochemical patterns. Iteration of this process enables 4D evolution of protein patterns within gels. b-e. Masked-based photolithographic techniques (λ = 365 nm) were utilized to control sequential protein patterning in defined shapes extending throughout the gel thickness. b. Following directed exposure of gels uniformly functionalized with mCherry-oNB-N3 (red), mCherry is released while uncaging sites for subsequent mCerulean-CHO (blue) immobilization by oxime ligation (c). d. A second round of directed light exposure released additional mCherry while leaving mCerulean patterns intact. e. EGFP-oNB-CHO (green) is immobilized in the uncaged alkoxyamine sites to create a trifunctional protein pattern. f-i. Evolution of trifunctional protein patterns was controlled in 3D space through multiphoton laser-scanning lithography (λ = 740 nm). Rows f-i correspond to 3D renderings of the photoevolved materials, where red channels represent minimum intensity projections and green and blue are maximum intensity projections, as well asxy, yz, and xz planar slices. f. Photorelease of mCherry-oNB-N3 is followed by (g) protein backfilling with EGFP-oNB-CHO. h. Further treatment with pulsed laser light released both EGFP-oNB-CHO and mCherry-oNB-N3 within user-specified gel sub-volumes while creating anchoring sites for (i) mCerulean-CHO immobilization. Images in b-i were generated using fluorescence confocal microscopy on a single gel throughout sequential patterning. Scale bars = 100 μm.