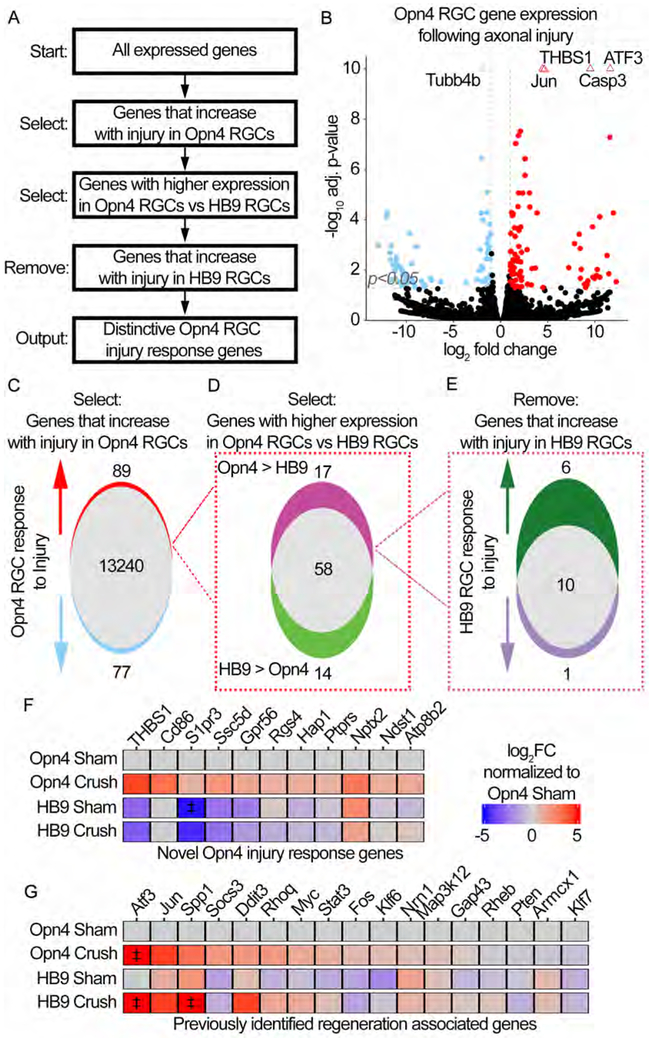
Figure 4: RNA-seq of regeneration competent ipRGCs reveals the expression of novel injury response genes.
(A) Multiple comparison methodology used to determine unique ipRGC injury response genes.
(B) Volcano plot showing differential gene expression in Opn4 RGCs following crush. Positive log2FC indicates an increase in Crush relative to Sham. Genes are considered significantly different if expression ±1 log2FC and adjusted (adj) p-value ≤0.05, vertical and horizontal reference lines at respective values. Triangles indicate genes with an adj p-value≤1*10−10, these values were fixed at 1*10−10.
(C) Venn-diagram representation of (B). 89 genes are upregulated following crush in Opn4 RGCs.
(D) Venn-diagram dividing the 89 genes from (C) based on the relative expression in injured Opn4 vs HB9 RGCs. 17 of the 89 genes are enriched in Opn4 RGCs.
(E) Venn-diagram dividing the 17 genes from (D) based on the expression change in HB9 RGCs following crush.
(F-G) Heatmaps showing the expression of indicated genes. Expression is presented as the log2FC relative to Sham Opn4 RGCs. ‡ indicates values that exceeded ±5 log2FC and were fixed at ±5 log2FC. (F) Heatmap of the 11 genes identified to be novel Opn4 injury response genes. (G) Heatmap of previously reported regeneration associated genes (RAGs).