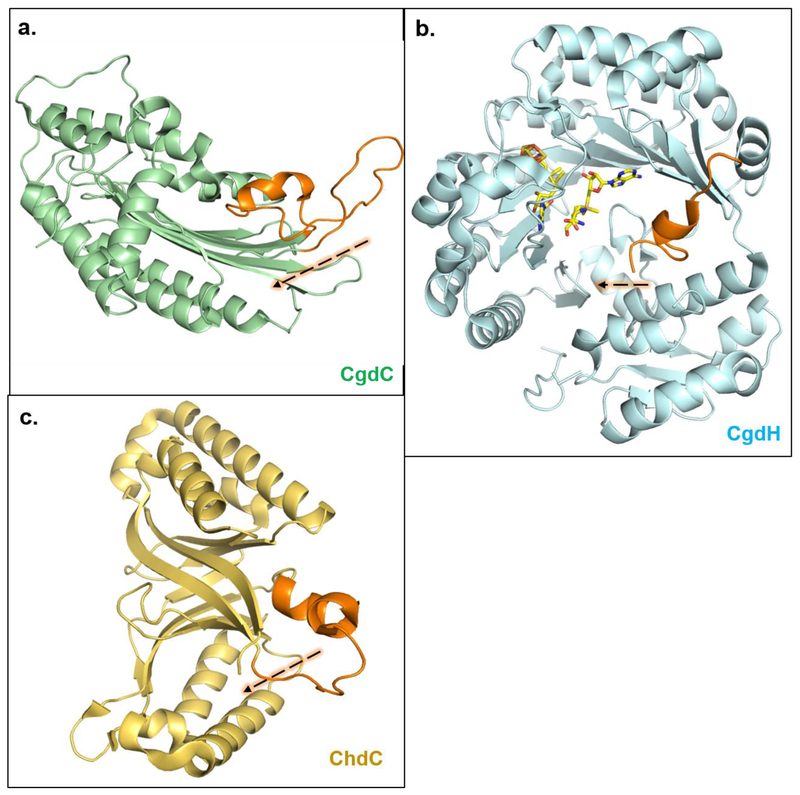
Figure 3. CgdC, CgdH, and ChdC all have an active site gate which closes upon substrate binding.
CgdC ((a); PDBID: 1TLB), CgdH ((b), PDBID: 1OLT) and ChdC ((c), PDBID: 1T0T) are isofunctional enzymes with no structural relationship and highly divergent reaction mechanisms. Interestingly, they all bind substrate at each monomeric site and structurally they share an “active site gate” (shown orange) made up of a partially disordered and mobile alpha helix, which closes in towards substrate upon its binding [4,9,11]. Note: Structures shown here are of individual monomers in their apo-/open- form. CgdC is a dimer, CgdH is a monomer, and ChdC is a pentamer in solution.