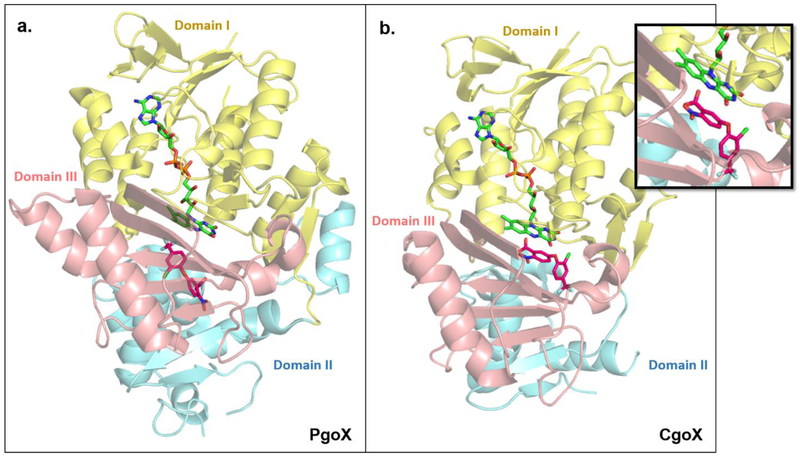
Figure 4. PgoX and CgoX share similar structural folds but show large differences in their active sites.
PgoX ((a); PDBID: 3NKS) and CgoX ((b); PDBID: 3I6D) share an overall folding pattern, consisting of three domains; (I) FAD-binding (yellow) domain, (II) membrane-binding domain (blue), and (III) substrate-binding domain (pink). Note: about 40% of amino acids in domain II of CgoX could not be crystallographically characterized [22]. PgoX and CgoX both non-covalently bind FAD (lime green) in the same location and orientation. The well-known PgoX inhibitor acifluoren (hot pink), also binds in both enzymes, but is mostly solvent-exposed in the CgoX active site, while it is buried deep inside the active site of PgoX. The different binding location and orientation of acifluoren in CgoX positions it to have a pi-stacking interaction with FAD (inset). This and the significantly larger active site in CgoX support the possibility that CgoX may use NAD(P) as an alternate electron acceptor to O2 to catalyze its reaction. Structures of PgdH1 and PgdH2 are not available to date.