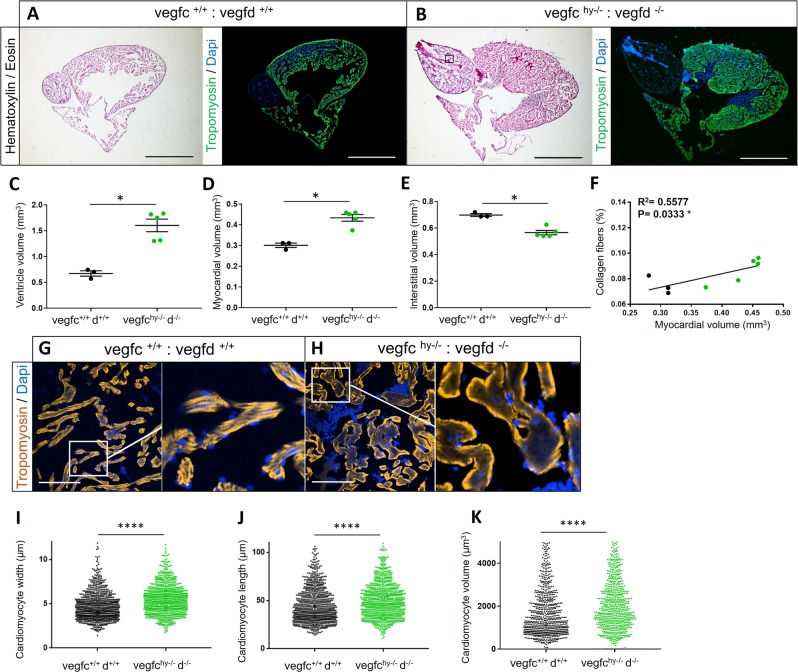
Fig. 3.
Loss of Vegfc/d signaling causes cardiac hypertrophy. Hematoxylin and Eosin or Tropomyosin and Dapi staining were performed on vegfc+/+:vegfd+/+ (a) and vegfc−/−:vegfdhy−/− hearts (b). Scale bar length: 0.8 mm. Quantification of ventricle volume (c), myocardial volume (d), and interstitial volume (e) in vegfc+/+:vegfd+/+ and vegfchy−/−:vegfd−/− mutant hearts. There is marked cardiac hypertrophy with a reduced interstitial volume in vegfchy−/−:vegfd−/− double mutants. n = 3 vegfc+/+:vegfd+/+ control hearts and 4 vegfchy−/−:vegfd−/− mutant hearts. Statistical analysis: unpaired, nonparametric, Mann–Whitney test was used to compare two means. Data are presented as the mean ± standard error of the mean (sem). f Correlation analysis between myocardial volume and collagen content in vegfc+/+:vegfd+/+ and vegfchy−/−:vegfd−/− mutant hearts. Black plots represent vegfc+/+:vegfd+/+ samples, green plots represent vegfchy−/−:vegfd−/− samples. There is a highly significant correlation between fibrosis and myocardial volume. g, h Tropomyosin and Dapi staining were performed on vegfc+/+:vegfd+/+ and vegfc−/−:vegfdhy−/− hearts. Scale bar length: 100 μm. i–k Enzymatic digestion was performed on vegfc+/+:vegfd+/+ or vegfchy−/−:vegfd−/− mutant ventricles and isolated cardiomyocytes were measured. The black dots represents the vegfc+/+:vegfd+/+cardiomyocytes width (i), length (j), or volume (k) and the green dots represent vegfchy−/−:vegfd−/− mutant cardiomyocytes width (i), length (j), or volume (k). There are statistically significant differences in cardiomyocyte size between control and mutant hearts. Frequency distribution calculated from a total of ~200 cardiomyocytes per samples (~800 cardiomyocytes per group) comprising n = 4 vegfchy−/−:vegfd−/− ventricles and n = 4 vegfc+/+:vegfd+/+ ventricles, respectively. Statistical analysis: unpaired, nonparametric, Mann–Whitney test was used to compare the two groups. Significant differences are marked by an asterisk ****p < 0.0001