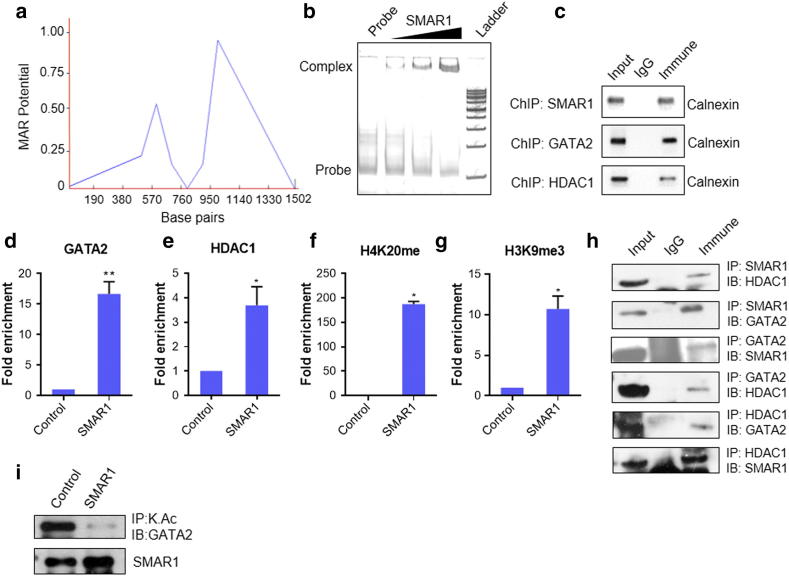
Figure 2.
SMAR1, GATA2 and HDAC1 binds to calnexin promoter. (a) MAR-WIZ software analysis to predict MAR site in calnexin promoter (b) Electrophoretic mobility shift assay (EMSA): 500 ng, 1 μg and 2 μg of SMAR1 DNA binding domain was used. MAR region spanning 100 bp was used as probe. (c) Chromatin immunoprecipitation (ChIP) assay was done for the binding of SMAR1, GATA2 and HDAC1 on calnexin promoter MAR region. Chromatin from HCT116 p53+/+ cells was immunoprecipitated with α-SMAR1, α-GATA2 and α-HDAC1 and quantitative real time PCR was performed and the relative fold enrichment of (d) GATA2 (e) HDAC1 (f) H4k20me and (g) H3k9me3 on calnexin gene promoters was determined. Parallel ChIP with α-mouse and α-rabbit IgG was used as control depending upon the isotype of antibody used for the immunoprecipitation. (h) Immunoprecipitation (IP), cell lysate from HCT116 p53+/+ cells was immunoprecipitated with α-SMAR1, α-GATA2 and α-HDAC1 antibodies (lane 3). Parallel immunoprecipitation with control rabbit IgG antibody is shown in lane 2. Lane 1 denotes input control. (i) Immunoprecipitation of adeno-SMAR1 transduced HCT116 p53+/+cells with acetyl lysine antibody and probed with GATA2 antibody. (Bars represent standard deviation from three independent experiments. */** indicate statistically significant difference at P ≤ .05/0.005, respectively).