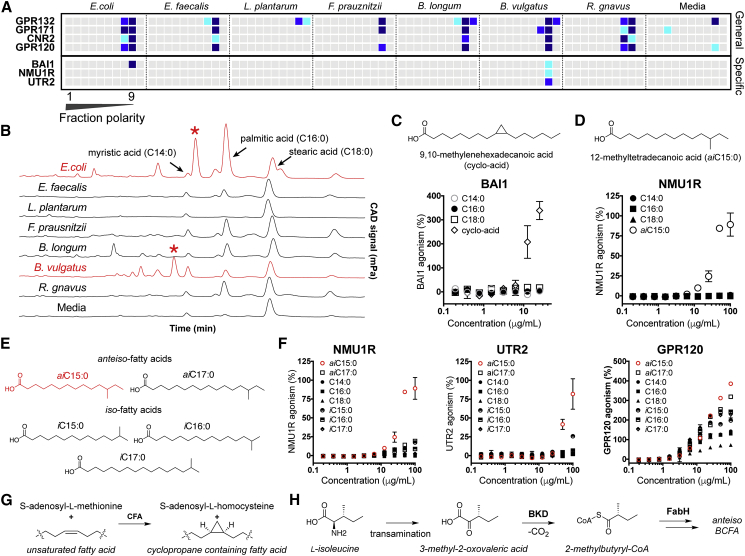
Figure 5.
Lipid Responsive GPCRs
(A) Heatmap of GPCRs demonstrating general (top) or specific (bottom) responses to lipid-rich fractions of bacterial extracts.
(B) Overlaid CAD chromatograms with common lipids and unique lipids (red asterisk) are marked.
(C) Structure of BAI1-active lipid 9,10-methylenehexadecanoic acid isolated from E. coli LF82, and the response of BAI1 to various fatty acids.
(D) Structure of NMU1R-active lipid, 12-methyltetradecanoic acid isolated from B. vulgatus, and the response of NMU1R GPCR to various fatty acids.
(E) Panel of branched chain fatty acids tested for GPCR fidelity.
(F) Response of NMU1R, UTR2 (specific), and GPR120 (general) to branched chain fatty acid panel.
(G) Biosynthesis of cyclopropane rings from unsaturated fatty acids using cyclopropane fatty acid synthase (CFA).
(H) Early steps in the biosynthetic scheme for ante-iso branched chain fatty acids (BCFAs) in bacteria (BKD, branched-chain α-keto acid dehydrogenase and FabH, β-ketoacyl-acyl carrier protein synthase III).
All dose-response curves were run in duplicate. Error bars are standard deviation. Error bars that are shorter than the height of the symbol are not shown.