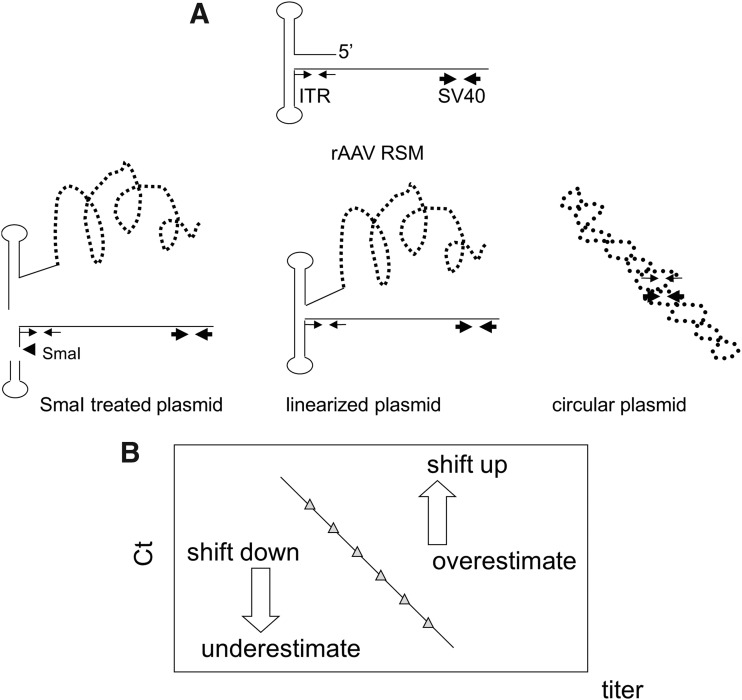
Figure 2.
The secondary structure of the plasmid DNA tail disturbed the amplification of ITR by qPCR. (A) The illustrations show the left part of the rAAV2RSM positive strand and the different secondary structures of pTR-UF11 plasmid DNAs. Thin arrows and thick arrows indicate the position of ITR and SV40 target sites, respectively. The arrowhead indicates the cutting site of SmaI. The treatment of pTR-UF11 with SmaI disrupted the ITR secondary structure. qPCR amplification was more efficient in the absence of hindrance by the plasmid tail, in SmaI-digested plasmid, and in the rAAV genome compared with linearized plasmid or circular plasmid. This resulted in up shift or down shift of the standard curves and thus overestimation or underestimation of the values measured by qPCR as shown in (B).