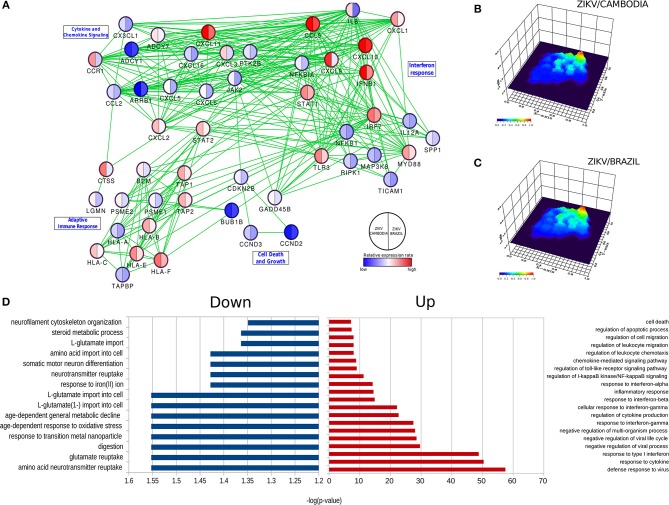
Figure 2.
ZIKV-dependent specific cellular processes induced in hiNPCs. (A) Specific networks induced by ZIKV/Brazil (right) and ZIKV/Cambodia (left) infected hiNPCs. Selected modules from the protein-protein interaction network were grouped according to KEGG pathways. Color gradient ranges from blue (down-regulated in infected cells) to red (up-regulated in infected cells), and it represents the mean fold change (log10) in expression levels relative to control (non- infected cells). (B) Gene network (from protein-protein interaction network) represented by a three-dimensional network topology view evidencing a strong upregulation of the interferon response in ZIKV/Brazil infected hiNPCs. (C) Gene network (from protein-protein interaction network) represented by a three-dimensional network topology view evidencing a strong upregulation of the interferon response in ZIKV/Cambodia infected hiNPCs. Color gradient represents the transcriptional activity of infected cells from blue (low activity) to red (high activity). (D) Gene Ontology (GO) enrichment analysis in “Biological Process” category statistically overrepresented among all differentially expressed genes. GO terms were represented by 2-fold enrichment value, with p-values <0.05. Infection experiments and control groups were performed in triplicate (n = 3).