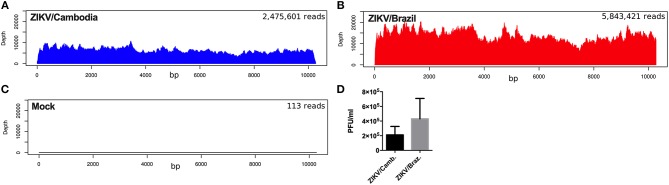
Figure 4.
ZIKV replication in hiNPCs. Sequencing results of the infected hiNPCs were mapped to the reference genome of Zika virus (x-axis). The depth of coverage achieved from infected hiNPCs cultures and mock (non-infected control cultures) was calculated for each condition as the total number of reads (average depth), mapped against a reference genome. ZIKV/Brazil infected hiNPCs reads were mapped against the reference genome: ZIKV/Brazil/PE243/2015 (GenBank: KX197192), ZIKV/Cambodia infected hiNPCs were mapped against the reference genome ZIKV/Cambodia/FSS13025/2010 (GenBank: JN860885.11). (A) Mock–non-infected cells. (B) hiNPCs infected with ZIKV/Brazil, analyzed at 48 h post infection. (C) hiNPCs infected with ZIKV/Cambodia, analyzed at 48 h post infection. (D) hiNPCs were infected in triplicate (n = 3) with either ZIKV/Brazil or ZIKV/Cambodia at a multiplicity of infection of 0.1. Infectious virus recovery in supernatant at 48 h post infection was determined by plaque titration assay. Values are expressed as mean of plaque forming units per milliliter (PFU/mL). Bars are representative of individual infection experiments performed in triplicate (n = 3).