Figure 1.
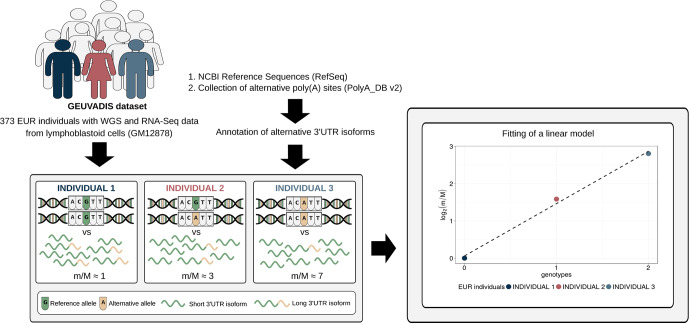
Schematic representation of the method. Genotypic data paired with RNA-Seq data from a large cohort of individuals are required to perform alternative polyadenylation quantitative trait loci (apaQTL) mapping analysis. RNA-Seq data are exploited, together with an annotation of alternative 3′ untranslated region (UTR) isoforms, to compute for each gene the m/M value that is proportional to the ratio between the expression of its short and long 3′ UTR isoforms. Then, the association between the m/M values of a gene and each nearby genetic variant is evaluated by linear regression. Genotypes are defined in the standard way: 0 means homozygous for the reference allele, 1 means heterozygous, and 2 indicates the presence of two copies of the alternative allele.