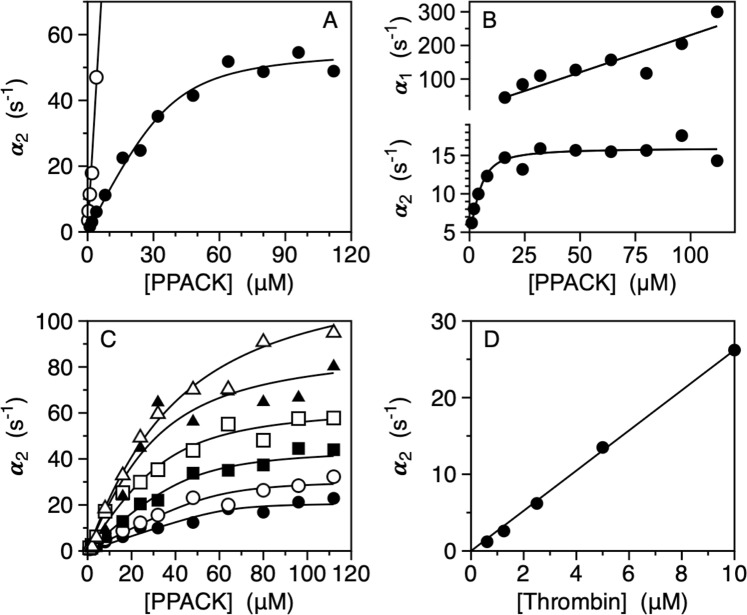
Figure 1.
(A) Rapid kinetics of PPACK binding to free thrombin (closed circles) showing a single, saturable relaxation that increases hyperbolically with the ligand concentration. The continuous line was drawn according to Eq. 2 in the text with best-fit parameter values listed in Table 1. In the presence of Na+, the kinetic profile changes into a straight line (open circles) that obeys the lock-and-key expression16,49 α = koff + kon[L], with best-fit parameter values koff = 0 s−1 and kon = 11 ± 1 μM−1s−1. (B) Rapid kinetics of PPACK binding to the thrombin mutant S195A showing the two relaxations predicted by the mechanism of conformational selection in Eq. 1. Continuous lines were drawn according to Eq. 2 in the text with best-fit parameter values listed in Table 1. (C) Rapid kinetics of PPACK binding to free thrombin over the temperature range 5–30 °C. The continuous lines were drawn according to Eq. 2 in the text with each rate constant expressed as in Eq. 3 and represent a global fit of the entire data set with best-fit parameter values: kon = 1.1 ± 0.1 μM−1s−1, k12 = 45 ± 3 s−1, k21 = 6.4 ± 0.4 s−1, Eon = 20 ± 3 kcal/mol, E12 = 12 ± 1 kcal/mol, E21 = 37 ± 5 kcal/mol. The values of the rate constants are at the reference temperature T0 = 288.15 K (15 °C) for the sake of comparison with the data in Figs 1A and 2. Experimental conditions are: 400 mM ChCl, 50 mM Tris, 0.1% PEG8000, pH 8.0 at 5 °C (closed circles), 10 °C (open circles), 15 °C (closed squares), 20 °C (open squares), 25 °C (closed triangles), 30 °C (open triangles). (D) Rapid kinetics of PPACK binding to thrombin in the presence of excess enzyme. The single, saturable hyperbolic increase observed with excess ligand (A) becomes a simple linear relaxation in the presence of excess macromolecule if the interaction takes place according to the mechanism of conformational selection. The slope of the straight line (2.1 ± 0.1 μM−1s−1) is a measure of the kon times the fraction of thrombin in the E form. The value is in good agreement with the best-fit values obtained from analysis of the data in panels A (Table 1) and C. Experimental conditions are: 400 mM ChCl, 50 mM Tris, 0.1% PEG8000, pH 8.0, at 15 °C. Data in the presence of Na+ (A) were obtained by replacing ChCl with NaCl in the buffer. Experimental errors for all data shown in Fig. 1A–D are 5% or less.