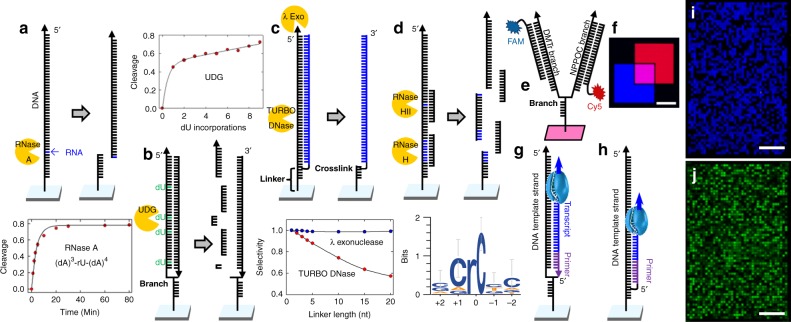
Fig. 4.
Post-synthetic enzymatic processing (Level-Three patterning). Nucleic acid sequences can be selectively or programmably cleaved, digested, or transcribed enzymatically on the biochip. a RNase A is a fast and efficient cleavage agent and requires only a single RNA nucleotide incorporation, optimally a pyrimidine surrounded by purines. b Similarly, uracil-DNA glycosylase (UDG) cleaves at pre-defined dU positions, but is significantly less efficient than RNase A cleavage. c Unmodified DNA can be digested by DNase I or lambda exonuclease. In the case of branched or crosslinked nucleic acid structures, lambda exonuclease digests only from a 5′ terminus and does not pass the junction, leaving the 3′ terminal DNA branch intact. DNase can be used if the second branch is not DNA and will not significantly digest a DNA surface linker shorter than a few nucleotides. d Programmable logical control of cleavage can be achieved with RNase H or HII; cleavage only occurs in conjunction with DNA hybridization to the cleavage site. RNase HII requires only a single RNA nucleotide incorporation at the desired cleavage site but has a preferred sequence context for maximum efficiency. e Branched structures synthesized either with asymmetric branching phosphoramidites or through crosslinking, can be effectively combined with enzymatic processing. f Fluorescence scans showing branched structure labeled by hybridization to either or both branches (200 µm scale bar). g Transcription of a DNA template using a branched structure. The primer is synthesized on the second branch using reverse phosphoramidites, or h attached by crosslinking. In either case the primer can be elongated as DNA or RNA using appropriate polymerases. i Fluorescence scan of branched structured synthesized via templated primer elongation using a DNA polymerase and fluorescein labeled NTPs. j Fluorescence scan of DNA strand synthesized enzymatically and hybridized after template degradation with UDG (200 µm scale bars). Source data are provided as a Source Data file