Fig. 4.
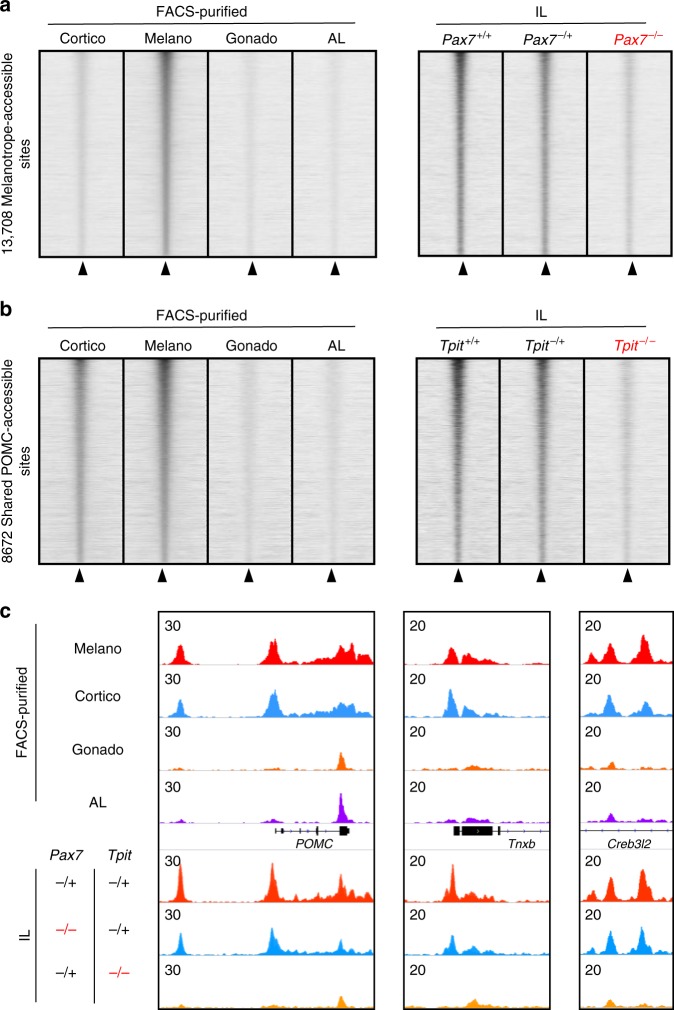
Pax7 and Tpit are required for opening cognate enhancer landscapes. a Read density heatmaps showing assay for transposase-accessible chromatin using sequencing (ATACseq) signals (reads per kilobase of transcript, per million mapped reads (RPKM)) across the different pituitary lineages in a 4 kb window centered at melanotrope-specific ATAC peaks (left panel). Right panel shows corresponding ATACseq heatmaps in the ILs of WT, Pax7+/−;Tpit+/− (labeled Pax7+/−) and Pax7−/−;Tpit+/− (labeled Pax7−/−) mice. b Read density heatmaps showing ATACseq signals (RPKM) across the different pituitary lineages in a 4 kb window centered at pro-opiomelanocortin (POMC)-specific ATAC peaks (left panel). Right panel shows corresponding ATACseq heatmaps in the intermediate lobe of WT, Pax7+/−;Tpit+/− (labeled Pax7+/−) and Pax7−/−;Tpit+/− (labeled Pax7−/−) mice. c Genome browser view (Integrative Genomics Viewer (IGV)) of ATACseq profiles at POMC-specific ATACseq peaks in the different pituitary lineages and in ILs of Pax7+/−;Tpit+/−, Pax7−/−;Tpit+/−, and Pax7+/−;Tpit−/− mice