Fig. 1. Distinct commensal microbial communities in anti–PD-1 responding patients and nonresponding patients as assessed with 16S rRNA gene amplicon sequencing.
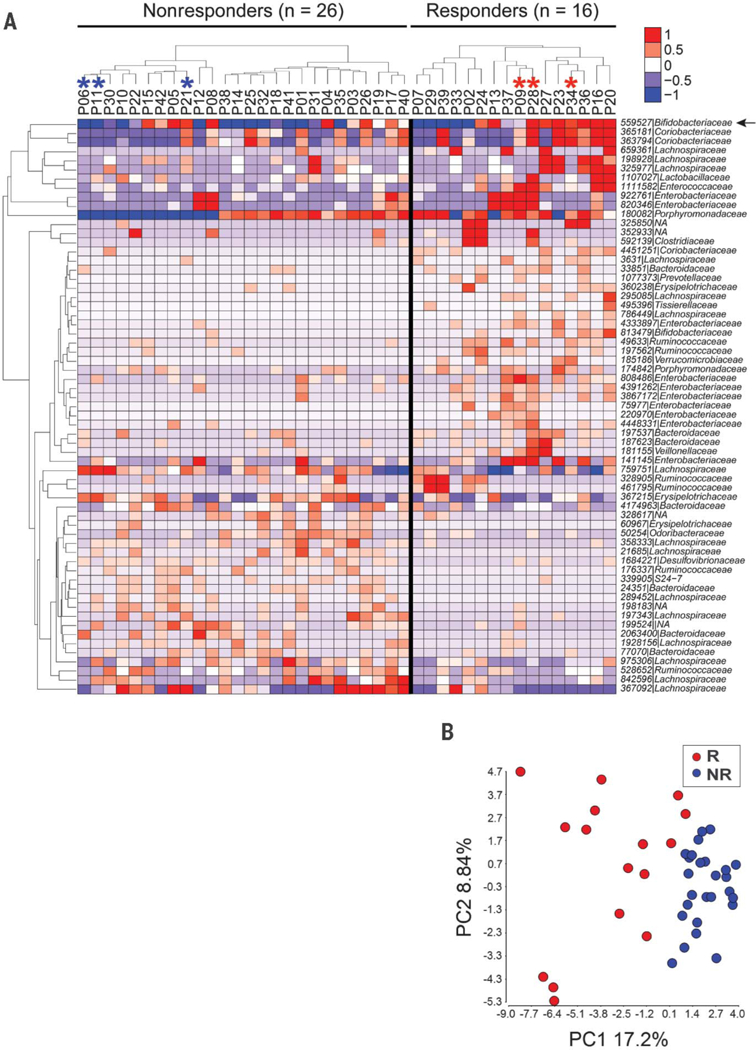
(A) Relative abundance of differentially abundant taxa in responders versus non-responders; 62 OTUs were identified as different with P < 0.05 (unadjusted, permutation test). An additional OTU 559527 (arrow) identified as Bifidobacteriaceae approached significance (P = 0.058). Hierarchical clustering of the samples was performed within each clinical group. Individual samples are organized in columns, labeled with patient identification number. Asterisks indicate samples used in further in vivo experiments. The ID of de novo assembled OTUs (new clean-up reference OTUs picked with QIIME) were abbreviated to show only the individual identifier digits, and the full OTU IDs are provided in table S4. (B) PCA of relative abundance of the 63 OTUs shown in Fig. 1A.
