Abstract
CRISPR-based technologies have become central to genome engineering. However, CRISPR-based editing strategies are dependent on the repair of DNA breaks via endogenous DNA repair mechanisms, which increases susceptibility to unwanted mutations. Here we complement Cas9 with a recombinase's functionality by fusing a hyperactive mutant resolvase from transposon Tn3, a member of serine recombinases, to a catalytically inactive Cas9, which we term integrase Cas9 (iCas9). We demonstrate iCas9 targets DNA deletion and integration. First, we validate iCas9's function in Saccharomyces cerevisiae using a genome-integrated reporter. Cooperative targeting by CRISPR RNAs at spacings of 22 or 40 bp enables iCas9-mediated recombination. Next, iCas9's ability to target DNA deletion and integration in human HEK293 cells is demonstrated using dual GFP–mCherry fluorescent reporter plasmid systems. Finally, we show that iCas9 is capable of targeting integration into a genomic reporter locus. We envision targeting and design concepts of iCas9 will contribute to genome engineering and synthetic biology.
Introduction
CRISPR and CRISPR-associated (Cas) systems, such as Cas9 nuclease and Cas12a (Cpf1) have drastically improved the ease of targeted DNA modifications. This is largely due to the ability to target Cas9's function via design and coexpression of single guide RNAs (sgRNAs) or CRISPR RNA for Cas12a.1–3 In the case of Cas9, sgRNA targeting is straightforward as it requires only simple DNA–RNA base pairing combined with the presence of a protospacer adjacent motif (PAM) on the target DNA.1,2 Systems employing Cas9 are highly robust and function in a broad range of organisms for a variety of editing strategies.4 Strategies for DNA integration and deletion are largely accomplished via formation of double-stranded DNA breaks (DSBs) or paired single-stranded DNA breaks followed by processing via endogenous nonhomologous end joining (NHEJ) or homologous recombination.4,5 Recently, groups have described homology-independent targeted integration an effective technique for NHEJ mediated genome integration.6–8 This technique produces simultaneous CRISPR-Cas9–targeted DSBs on plasmid and genomic protospacer sequences followed by NHEJ to ligate cleaved plasmid DNA into the genomic protospacer. However, it has become apparent that CRISPR-based genome engineering strategies are limited with respect to their dependence on the generation of DSBs and endogenous DNA repair machinery.9–11 DSBs could generate unwanted mutations, translocations, and complex rearrangements and destabilize karyotype.12,13 This is a fundamental limitation of CRISPR-Cas9's application in editing human cell lines and has led to the development of technologies that avoid incurring double-stranded DNA damages during the editing process. For instance, “base-editor” (BE) Cas9 systems enable generation of single nucleotide changes without the need for double-stranded DNA breaks.14,15 BE-Cas9 systems accomplish single nucleotide changes via fusion of a nicking Cas9 (Cas9D10A) with a cytidine deaminase and uracil glycosylase inhibitor domains. However, BEs are limited to single nucleotide changes.
Site-specific recombinases are also powerful tools for genome engineering and synthetic biology.16,17 These proteins are capable of facilitating DNA rearrangements with high predictability and specificity without incurring unwanted DSBs. Site-specific recombinases possess the enzymatic machinery to facilitate transient DNA cleavage, strand exchange, and religation without the need for high energy cofactors, DNA replication or cellular DSB repair.18 Integrases, such as ΦC31, are relatively limited to specific ∼30 bp recognition sites and are often used for integration at specific “landing pad” or pseudo-site loci.19,20 To circumvent this, directed evolution has been employed to retarget recombinase substrate specificity. For instance, Karpinski et al. reported directed evolution of Cre recombinase to target conserved sequences human immunodeficiency virus long terminal repeats. This system led to efficient and highly specific excision of the human immunodeficiency virus provirus. However, nearly 150 rounds of directed evolution were required.21 Alternatively, recombinases have been retargeted by fusing catalytic-domains to zinc finger or transcription activator–like DNA-binding domains.22,23 These techniques however require complex addition of heterologous DNA-binding domains. More recently a recombinase–Cas9 (recCas9) was created via fusion of dCas9 with Gin invertase.24 This system was able to target DNA deletion; however, it was unable to target DNA integration. In addition, recCas9 displayed low efficiencies when targeting genomic DNA.
To overcome the limitations with current Cas9-based site-specific recombinases, we investigated the development of Cas9 fusions that employ catalytically inactive Cas9 (dCas9) with a catalytic domain of a resolvase. Previous reports have generated heterologous fusions between resolvase catalytic and zinc finger DNA-binding domains. These reports have focused on employing recombinases, such as Gin invertase and Tn3 resolvase.22,25 This family of serine recombinase proteins benefit from compact and highly separated DNA-binding and catalytic domains compared to some tyrosine recombinases, such as Cre and FLP.18 We focused on Tn3 resolvase since there is extensive in vitro characterization of its hyperactivating mutations.26,27 Specifically, three-dimensional (3-D) protein structures for its DNA-binding (dimer) and DNA-synapsis (strand-exchange, tetramer) states support the potential of resolvase reengineering.28,29 In this study, we generated an RNA-guided recombinase through the fusion of dCas9 with the catalytic domain of Tn3 resolvase, herein referred to as integrase Cas9 (iCas9). We first demonstrate iCas9 functionality using a chromosomally integrated fluorescent reporter in yeast (Saccharomyces cerevisiae). Next, we demonstrate that in human cells (HEK293) iCas9 can facilitate plasmid-to-plasmid deletion and integration. Finally, we show that iCas9 allows for targeted integration into endogenous genomic loci. In the future, iCas9 will greatly expand the utility and fidelity of Cas9-based genome editing in downstream applications in synthetic biology, disease modeling, and regenerative medicine.
Methods
Bacterial culture
Molecular cloning was conducted using Escherichia coli NEB-10-Beta (New England Biolabs [NEB]). LB Miller medium (Sigma Aldrich, Sigma) was supplemented with appropriate antibiotics for plasmid maintenance: ampicillin (100 μg/mL) or chloramphenicol (30 μg/mL). E. coli were cultured at 37°C.
Yeast culture
All yeast were cultured at 30°C. S. cerevisiae YPH500 were propagated on YPD agar plates and in liquid medium containing glucose. Liquid cultures were shaken at 250–300 rpm. Yeast minimal dropout media contained either 2% glucose or 2% galactose with 1% raffinose and necessary amino acid dropout solutions (Clonetech). Yeast were made competent using the Zymo competent yeast kit (Zymo Research) and transformed using the manufacturer's protocol. Genomic integrations and plasmid transformations were selected for on yeast minimal dropout plates with amino acid combinations necessary for selection. Yeast were cultured in liquid yeast dropout media necessary for plasmid selection.
Mammalian cell culture
HEK293T cells (ATCC CRL-3216) were cultured on poly-L-ornithine (PLO) (Sigma) coated plates and maintained in Dulbecco's modified eagle medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin–streptomycin (all from ThermoFisher). Cells were maintained in a 37°C incubator with 5% CO2 and passaged once ∼80% confluent.
Molecular cloning
iCas9 (Tn3-GGS × 6-dCas9) was constructed by fusion of a previously described hyperactive mutant recombinase (Tn3G79S,D102Y,E124Q). The resolvase catalytic domain (AA1-148) was linked to Cas9D10A,H840A with a flexible glycine serine (GGS × 6) linker. N- and C-terminal SV40 nuclear localization sequences with small glycine serine linkers (GGS × 1) were added to facilitate nuclear entry. The coding region for the hyperactive Tn3 mutant resolvase was synthesized as a human codon optimized gBlock by Integrated DNA Technologies. The gBlock was subcloned into a dCas9 derivative of p415 Gal1-Cas9 (Addgene 43804).30 The mTn3 catalytic domain along with D10A and H840A mutations to Cas9 were added using PCR primers containing SapI sites (Supplementary Table S2). Purified PCR products were digested with SapI and gel-extracted using the Sigma-Aldrich gel-extraction kit. iCas9 was assembled in XbaI-XhoI sites of p415 Gal1-Cas9. The resulting p415 Gal1-iCas9 vector also contains a Cen6 origin of replication and a leucine prototrophic marker. For expression in human cells iCas9 was PCRed with primers adding AgeI and MfeI upstream and downstream respectively. iCas9 was cloned into a modified pX330 with guide expression cassette removed. Digested and gel-extracted iCas9 PCR products were ligated with AgeI and EcoRI digested pX330. The resulting vector contains a cytomegalovirus β-actin hybrid promoter driving iCas9 expression.
Single guide RNA guides were synthesized as pairs of oligonucleotides. Terminal 5′ phosphates were added to oligonucleotides by incubating 1μg total of top/bottom oligonucleotides in 50 μL reactions containing 1 × T4 DNA ligase buffer and 10 units of T4 polynucleotide kinase (T4 PNK) at 37°C overnight (Supplementary Tables S1 and S2). Oligonucleotides were duplexed by heating the kinase reactions to 90°C on an aluminum heating block for 5 min followed by slowly returning the reaction to room temperature (25°C) over approximately 1 h. Following duplexing, guides were ligated into respective vectors.
Yeast sgRNA expression cassettes were constructed by cloning oligonucleotide duplexes into, pSB1C3 containing an SNR52 promoter with inverted SapI sites and an sgRNA hairpin recognized by S. pyogenes Cas9. Pairs of sgRNAs were then PCRed with primers adding EcoRI and SapI or SapI and SpeI sites. Purified PCR products were then digested with respective restriction enzymes, heat inactivated, and ligated into EcoRI and SpeI digested pRS424.31 The resulting vector contains pairs of yeast sgRNA cassettes with a 2 μ origin of replication and tryptophan prototrophic marker.
Humanized sgRNAs were cloned into a modified pSB1C3 vector containing a human U6 promoter, inverted BbsI sites, and a Streptococcus pyogenes–recognized sgRNA hairpin (sequence derived from pX330). Pairs of sgRNAs were then PCRed with primers adding EcoRI and SapI or SapI and XbaI sites. Purified PCR products were then digested with respective restriction enzymes, heat inactivated, and ligated into EcoRI and XbaI digested pUC19. The resulting vector contains pairs of human sgRNA expression cassettes.
The yeast genomic integration vector (pMG) was generated using vectors previously described by Ellis and colleagues.32 Tef1 promoters drive constitutive expression of GFP and mCherry. To integrate into the yeast genome, 1–2 μg of pMG was digested with ApaI in 50 μL reactions for ≥1 hour at 37°C. Five microliters of restriction product was transformed into competent YPH500 using the Zymo Competent Yeast Kit (Zymo) following manufacturer protocols. Integrants were selected for by plating on histidine dropout plates.
To clone iCas9-target sequences into pMG, sites were synthesized as overlapping oligonucleotides. Terminal 5′ phosphates were added to oligonucleotides by incubating 1μg of top/bottom oligonucleotides in 50 μL reactions containing 1 × T4 DNA Ligase Buffer and 10 units of T4 polynucleotide kinase (T4 PNK) at 37°C overnight. Oligonucleotides were duplexed by heating the kinase reactions to 90°C on an aluminum heating block for 5 min followed by slowly returning the reaction to room temperature (25°C) over approximately 1 h. Following duplexing, sites were ligated into EcorI and MluI sites surrounding GFP.
Mammalian cell transfections
HEK293T cells were seeded at 1.8 × 105 cells/well in PLO coated 24-well plate and transfected 24 hours postpassage at ∼80% confluency. For plasmid–plasmid assays, 300 ng of iCas9, 100 ng of GFP-encoding donor vector (FeGFP-1C3), 100 ng of mCherry-expressing target vector (pUC:EAMP), and 100 ng sgRNA expression vectors were transfected per well using 1.5 μL Lipofectamine 3000 and 1 μL P3000. For genome integration experiments, 300 ng iCas9 expression vector, 100 ng GFP-encoding donor vector (FeGFP-1C3), 100 ng pIRFP670, and 100 ng sgRNA cassette(s) were transfected using 1.5 μL Lipofectamine 3000 and 1 μL P3000. pIRFP670 was cotransfected as a control with samples at >50% transfection efficiency.
Stable cell line generation
HEK293T cells were passaged to four PLO-coated 100 mm culture plates in Opti-MEM reduced serum medium plus GlutaMAX and supplemented with 1 mM sodium pyruvate and 10% (v/v) fetal bovine serum (all from ThermoFisher). To generate recombinant HEK293T, cells were transfected with the pKSBRV-1 transgene and packaging plasmids (pUMVC and pVSVG). Nine micrograms of pKSBRV-1, 6 μg pUMVC, and 3 μg pVSVG expression plasmids were transfected per plate using 28 μL Lipofectamine 3000 and 36 μL P3000 (ThermoFisher). Media was changed 6 h post-transfection and supernatant was collected at 24 h and 54 h. Conditioned media was filtered using 0.45 μm filter and particles were concentrated using Lenti-X (Takara Bio). HEK293T cells were transduced by incubation with supernatant media followed by puromycin selection 48 h later at a concentration of 0.75 μg/mL. Following selection for 2 weeks, cells were sorted using fluorescence activated cell sorting for the upper 50% of mCherry expressing cells to generate a pure population of cells stably expressing the transgene.
In yeast GFP deletion assay
To assay iCas9 function, YPH500 Ura3(MGaa) with p415 Gal1-iCas9 and with various pRS424 (guide pairs) were cultured in 3 mL YP –Leu, –Trp with 2% glucose. After 24 h, 5 μL of the stationary phase culture was used to inoculate 3 mL of YP –Leu, –Trp with 2% galactose, 1% raffinose. Cells were diluted down (5 μL saturated culture in 3 mL media) at 48 h intervals. Cells were analyzed by flow cytometry and fluorescent microscopy after 96 h of galactose induction. Genomic DNA was also prepared after galactose induction.
Flow cytometry
All flow cytometry was conducted on an Accuri C6 Flow Cytometer (BD Biosciences). Samples were gated by consistent forward scatter (FSC) and side scatter (SSC) and 10,000 events within the FSC/SSC gate were collected. A 488 nm laser excitation and a 530 ± 15 nm emission filter was used for GFP fluorescence determination. Flow cytometry files were analyzed using manufacture software and in MatLab (The MathWorks). Flow cytometry of HEK293T cells was conducted 72 h post-transfection. Briefly, cells were dissociated using Accutase (ThermoFisher), washed with phsophate-buffered saline (PBS), and analyzed using a BD Accuri C6 cytometer (BD Biosciences). GFP-positive cells were measured compared with transfections with a non-target sgRNA.
Fluorescent microscopy
Two hundred microliters of stationary phase cultures of yeast were spun down at 4000 g for 2 min and washed once in 1 × PBS solution. Cells were concentrated in 10–20 μL of 1 × PBS. To visualize cells, 1–2 μL of cell solution was placed on glass microscope slides and visualized on a Nikon Ti-Eclipse inverted microscope with an LED-based Lumencor SOLA SE Light Engine with appropriate filter sets. GFP was visualized with an excitation at 472 nm and emission at 520/35 nm using a Semrock band pass filter. mCherry was visualized with excitation at 562 nm and emission at 641/75 nm. Constant exposure times and image gain adjustments were applied to microscopy data. HEK293T cells were imaged directly on tissue-coated plates 72 h after transfection.
Genomic DNA isolation and PCR analysis of GFP deletions
Yeast genomic DNA was prepared using the Zymo yeast genomic DNA preparation kit per the manufacturer's protocol with phenonl/chloroform steps included. To assay genomic deletion, PCR was conducted using Phusion DNA polymerase (New England Biolabs). Annealing temperatures and extension times were calculated using the manufacturer's protocol. PCR products were visualized via 0.8% agarose gel electrophoresis. Human cell genomic DNA was prepared 72 hours post-transfection using the Qiagen DNEASY kit using the manufacturer protocol. PCR was conducted on 250 ng of genomic DNA with primers target the integration junction. Products were resolved on a 2% agarose gel.
Sequencing of deletion and integration products
Following gel resolution, PCR-derived amplicons of integration or deletion products were gel-extracted using the Gen Elute gel extraction kit (Sigma-Aldrich) per the manufacturer's protocol. Following extraction, products with phosphorylated via incubation in 50 μL reactions with T4 PNK and 1 × T4 DNA ligase buffer. Reactions were heat inactivated and ligated in equimolar ratio to SmaI cleaved and dephosphorylated pUC19. Ligations were transformed into chemically competent NEB10β E. coli and plated on ampicillin plates supplemented with 40 μL X-Gal solution (Promega). White colonies were picked and prepared using GeneElute Plasmid Preperation kit (Sigma-Aldrich). Three hundred nanograms of plasmid DNA was sequenced via DNASU's Sanger Sequencing Core facility.
Statistical analysis
Data are presented as mean of triplicate ± standard deviation. Statistical analyses included one-way ANOVA with Dunnett's multiple comparison test for genomic GFP deletion and integration. One-way ANOVA with Tukey's multiple comparison test was employed for plasma deletion. Student's one-tailed t-test was employed for plasmid-to-plasmid recombination. Interdomain linker analysis was performed uses a two-way ANOVA with Tukey's multiple comparison test. Analyses were performed in Graph Pad's Prism software.
Results
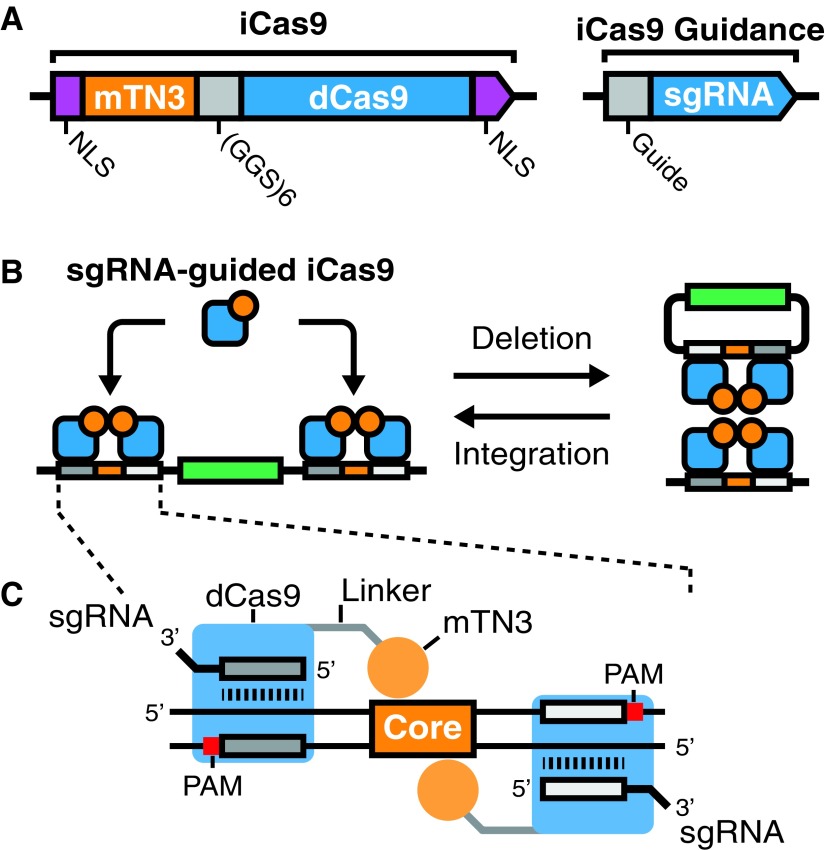
The design of iCas9 followed several general principles. First, the fusion of catalytically inactive Cas9 (dCas9) with a hyperactive mutant Tn3 resolvase (mTn3) was accomplished by addition of the N-terminal resolvase catalytic domain to the N-terminus of dCas9 (Fig. 1A). These domains were separated by a flexible glycine serine (GGS × 6) linker. To facilitate nuclear entry, we included SV40 nuclear localization sequences on both the N- and C-termini. The choice of mTn3 was motivated by previous studies that showed mTn3 zinc finger fusions were capable of DNA deletion and integration (Fig. 1B). Finally, previous work demonstrated FokI-dCas9 fusion proteins dimerize when pairs of sgRNAs were targeted in a PAM-distal orientation.33 This suggested that mTn3's N-terminal heterologous fusion with dCas9 are presented adjacent to the 5′ end of the sgRNA bound to a protospacer DNA. Furthermore, solved protein structures for streptococcus pyogenes Cas9 place the N-terminus closer to the 5′ end of the sgRNA than the C-terminus.34 Collectively, structural information and previous FokI-dCas9 results strongly suggest that a PAM-distal protospacer orientation flanking a mTn3 core recognition site should enable RNA-guided targeting (Fig. 1C).
FIG. 1.
Design of integrase CRISPR-associated protein-9 nuclease (iCas9) and iCas9 target sites. (A) Architecture of the iCas9 fusion protein. A catalytically inactive Cas9 (dCas9, blue) is fused to the catalytic domain of a hyperactive mutant recombinase from transposon TN3 (mTN3, orange). dCas9 and mTN3 are separated by a flexible linker region (GGS × 6, gray). To promote nuclear entry, both N- and C-termini have SV40 nuclear localization signals sequences (NLS, purple). Given catalytic domains are fused to dCas9, iCas9 is guided via single guide RNAs (sgRNAs, blue). (B) mTN3 function is dependent on dimerization on target site sequences, followed by tetramerization. Tetramerization results in recombination, which can occur in two directions: deletion or integration. iCas9 can target either DNA deletion if target recognition sites are located on the same molecule (left), or alternatively iCas9 can target DNA integration if target sites are on separate DNAs (right). (C) The design of an iCas9 recognition site consists of two sgRNA targets (dark and light gray) flanking a TN3 Res1 core recognition sequence (core, orange). The two sgRNAs have a protospacer adjacent motif (PAM, red) distal orientation. mTN3–dCas9 fusions bind in positions around the core sequence allowing for mTN3 catalytic domain dimerization.
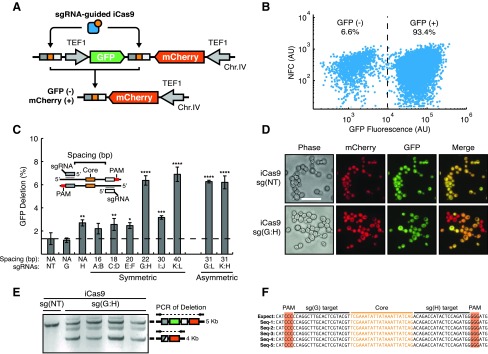
To demonstrate that iCas9 targets eukaryotic genomic DNA, we constructed a Saccharomyces cerevisaea dual-fluorescent recombination reporter system, which contains GFP and mCherry expression cassettes and enabled detection of recombination using flow cytometry and fluorescence microscopy.35 Both GFP and mCherry were constitutively expressed from translation elongation factor 1 (Tef1) promoters. The GFP cassette was flanked by Tn3 Res1 core sequences and resulted in GFP deletion upon iCas9 targeting (Fig. 2A, Supplementary Fig. S1). Each core sequence was flanked with numerous PAMs, which enabled systematic analysis of sgRNA spacings (Table 1, Supplementary Fig. S2). iCas9 was placed on a yeast Cen6 vector with galactose inducible promoter, and sgRNAs were placed on a yeast 2 μ vector with SNR52 promoters (Supplementary Fig. S1). We found coexpression of iCas9 along with targeting sgRNA pairs resulted in loss of GFP detectable by flow cytometry (Fig. 2B). Expression of a nontarget sgRNA [sg(NT)] or single sgRNA did not result in statistically significant GFP deletion, however sg(H) did result in subtle levels of GFP deletion (Fig. 2C). This is consistent with previous reports indicating that hyperactive recombinases are capable of spontaneous dimerization and recombination.36 We hypothesized that mTn3's dimerization-dependent function would preferentially operative via cooperative targeting by sgRNAs. As such, we systematically analyzed sgRNA spacing's from 16 bp to 40 bp. Consistent with this, we found symmetric spacings of 22 [sg(G:H)] and 40 bp [sg(K:L)] resulted in 6.4 ± 0.4% and 6.9 ± 0.6% GFP deletion, respectively. However, 30 bp spacing symmetrically [sg(I:J)] placed around the core sequence reduced the GFP deletion efficiency. Alternatively, asymmetric spacings of 31 bp [sg(G:L) or sg(K:H)] around the core resulted in GFP deletion efficiencies similar to those observed for symmetric spacings of 22 and 40 bp (Fig. 2C). Interestingly, the observed functional spacings are consistent with the requirement for targeting resolvase monomers to the same DNA helical face (see Supplementary Fig. S3).
FIG. 2.
Validation of iCas9 function and target site design using a yeast-based GFP-deletion assay. (A) A diagram of chromosomally integrated dual-fluorescent reporter for detection of iCas9 function. The reporter contains GFP (green) and mCherry (red) coding regions transcribed from separate TEF1 promoters (gray arrows). iCas9 recognition sites flank the GFP expression cassette, wherein each site contains a left (dark gray rectangle) and right (light gray rectangle) protospacers flanking a TN3 Res1 core sequence (orange square). Functional targeting of iCas9 results in GFP deletion generating GFP−, mCherry+ cells. (B) A representative flow cytometry scatter plot for yeast expressing the reporter, iCas9, sgRNAs G and H after 96 h of galactose induction of iCas9 expression. NFC, nonfluorescent channel. (C) Systematic analysis of sgRNA spacing on iCas9 function, as measured by GFP deletion on flow cytometry. Inset shows spacing as measured from the 5′ ends of sgRNAs flanking the core sequence. sgRNAs A–M are systematically spaced around the core sequence and distances ranging from 16 to 40 bp. Nontarget sgRNA [sg(NT)] is a control guide not matching the target site, where the dashed line indicates background false GFP deletion. “Symmetric” indicates left and right guides are positioned equal distances around the core site. “Asymmetric” guide combinations are at varying distances from the core. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared with sg(NT) by one-way ANOVA with Dunnett's multiple comparison test. (D) Fluorescent microscopy of yeast expressing iCas9 and sg(NT) or the 22 bp targeting pair, sg(G:H). GFP and mCherry dual-positive cells are orange on merge, while GFP deletions appear as red only (GFP−, mCherry+). Scale bar is 20 μm. (E) Gel electrophoresis of amplicons using primers flanking the reporter locus. The starting reporter results in a 5 kilobase (kb) PCR product and GFP deletion results in a 4 kb amplicon. Coexpression of iCas9 and sg(G:H) results in detectable DNA deletion via formation of the 4 kb product. (F) Sequencing of iCas9 target sites from isolated and subcloned deletion amplicons. Sequencing results (Seq1–Seq5) aligned to the expected recombination product (Expect). Deletion products match the expected recombination sequence and are free of insertion deletion (indel) mutations.
Table 1.
Single guide RNA guide sequences and iCas9-site design
| sgRNA | Position | Spacing (bp) | Guide sequence |
|---|---|---|---|
| A | Left | 16 | CGAACGTACGAGTGCAAGCC |
| B | Right | 16 | CAGACAGACCATACTCCAGA |
| C | Left | 18 | GAACGTACGAGTGCAAGCCT |
| D | Right | 18 | AGACAGACCATACTCCAGAT |
| E | Left | 20 | AACGTACGAGTGCAAGCCTG |
| F | Right | 20 | GACAGACCATACTCCAGATG |
| G | Left | 22 | ACGTACGAGTGCAAGCCTGG |
| H | Right | 22 | ACAGACCATACTCCAGATGG |
| I | Left | 30 | ACGAGTGCAAGCCTGGGGGA |
| J | Right | 30 | ACCATACTCCAGATGGGGGA |
| K | Left | 40 | TGCAAGCCTGGGGGATGGAT |
| L | Right | 40 | ACTCCAGATGGGGGATGGCT |
| NT | NA | NA | AGAAGAGCGAGCTCTTCT |
| iCas9 site | ||
|---|---|---|
| Left | 20 Bp Core | Right |
| TCCGATCCATCCCCCAGGCTTGCACTCGTACGTTCGAAATATTATAAATTATCAGACAGACCATACTCCAGATGGGGGATGGCTAGGT | ||
Single guide RNA (sgRNA) names (A–L) are indicated with position relative to the core sequence (left/right). Base pair (bp) spacings between tested sgRNA pairs (Fig. 1C) are indicated. Specific guide and iCas9-site nucleotide sequences are indicated. The 20 bp TN3 core sequence is underlined. NT, nontarget control sgRNA; NA, not applicable.
To confirm that loss of GFP was due to GFP deletion and not the result of spurious cell death, we utilized fluorescence microscopy to detect GFP and mCherry expression. We found all cells with a nontarget guide [sg(NT)] did not result in GFP deletion as all cells expressed both GFP and mCherry. However, cooperative targeting with sgRNA pairs (sg[G:H]) resulted in GFP-negative cells with intact mCherry expression (Fig. 2D). To verify that recombination occurred on the endogenous DNA level, we performed PCR with primers flanking the GFP and mCherry expression cassettes. In the absence of GFP deletion, the PCR reaction would result in a 5 kb product while GFP deletion would result in a 4 kb amplicon. As expected, we observed the formation of this 4 kb deletion-dependent product when we coexpressed iCas9 with sgRNA pairs [sg(G:H)] but did not observe this product when iCas9 was employed with a nontargeting sgRNA [sg(NT)] (Fig. 2E). To further characterize the deletion product, we isolated, subcloned, and Sanger sequenced the 4 kb deletion amplicons and observed the expected recombination product without insertion deletion (indel) mutations (Fig. 2F). Together, these results indicate that iCas9 targets DNA deletion and its function is dependent on RNA guidance.
Aiming to improve iCas9 function, we speculated that the interdomain linker amino acid sequences in our iCas9 domains could improve DNA targeting efficiency. To that end, we engineering several iCas9 variants with a range of flexible glycine serine and rigid linkers, including those that have been previously used with Cas9 heterologous fusions proteins.15,33 We assessed the efficiency of these iCas9 variants in our GFP deletion assay with two sets of sgRNA pairs [sg(G:H) and sg(K:L)]. We did not observe a marked increase in GFP deletion efficiency with these other iCas9 variants versus our original iCas9 construct, which employed a flexible glycine serine (GGS × 6) linker (Supplementary Fig. S1). Therefore, we proceeded with subsequent evaluation of iCas9 using the (GGS × 6) linker design.
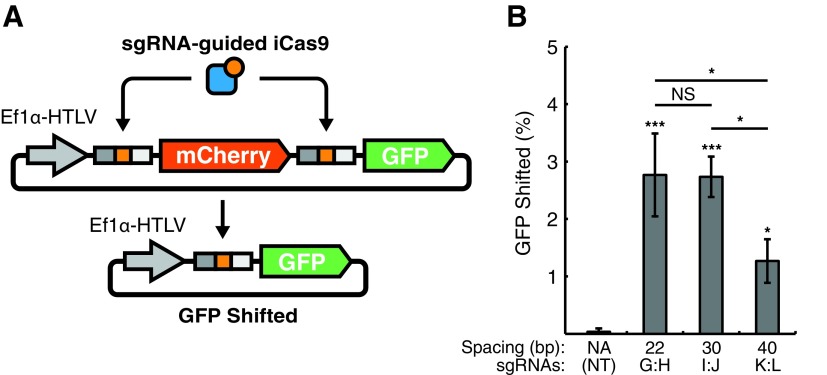
To assess the function of iCas9 in human cells, we developed a dual-fluorescence detection plasmid-based reporter. The reporter plasmid contained mCherry flanked by core recognition sites with GFP downstream (Fig. 3A and Supplementary Fig. S5A). Therefore, mCherry deletion would result in cells expressing GFP only, yielding a population of cells with GFP levels shifted over mCherry. We cotransfected HEK293T cells with dual-reporter, sgRNA, and iCas9 expression vectors while gating out untransfected cells. The shift of cells with GFP over mCherry expression was quantified using flow cytometry to evaluate sgRNA spacings for our plasmid targeting assay. While the use of a nontargeting sgRNA [sg(NT)] did not result in a GFP shift, sgRNA pairs with spacings of 22 [sg(G:H)], 30 [sg(I:J)], and 40 bp [sg(K:L)] resulted in a statistically significant increase in shifted GFP (Fig. 3B). Moreover, these results indicated that sgRNA pairs with 22 [sg(G:H)] and 30 bp [sg(I:J)] spacings were comparably functional (Fig. 3B). These results are interesting because previous work with Gin-dCas9 fusions have reported the ability for 30 bp sgRNA spacing to target DNA deletion on plasmid substrates.24 This may be due to the use of supercoiled plasmids as substrates, which may support less stringent spacing requirements due to DNA coiling and 3-D presentation. Nevertheless, because 22 bp sgRNA spacing worked efficiently in both plasmid and genomic assays, we chose to proceed with this sgRNA pair for subsequent evaluation of iCas9.
FIG. 3.
Detection of iCas9 function using an episomal deletion assay in human cells. (A) A dual-fluorescence plasmid systems contains an EF1α-HTLV promoter (gray arrow), iCas9 recognition sites (gray and orange rectangles) flanking mCherry (red), and a downstream GFP (green) reading frame. iCas9 targeting results in deletion of mCherry and generation of a GFP-only vector. (B) HEK293Ts were cotransfected with the GFP–mCherry reporter plasmid, iCas9, and guides targeting the recognition sites. Cotransfection of iCas9 and a sg(NT) resulted in no shift of GFP expression. However, targeting with 22, 30, and 40 bp sgRNA spacings spacing shifted GFP by 2.8 ± 0.7, 2.7 ± .0.4 and 1.3 ± 0.4 % respectively. *p < 0.05, **p < 0.01, ***p < 0.001, by one-way ANOVA with Tukey's multiple comparison test. NS, nonsignificant.
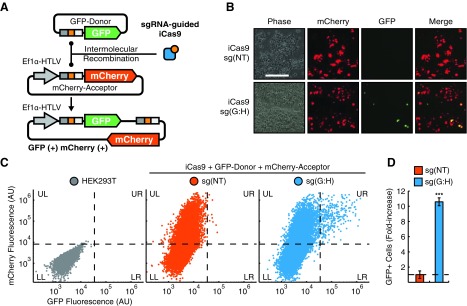
Next, to determine iCas9's ability to target intermolecular recombination, we developed a two-plasmid reporter system for plasmid-to-plasmid integration. In this system, one plasmid contains an elongation factor 1α (EF1α) human T-cell leukemia virus (HTLV) hybrid promoter, and a core target site upstream of a mCherry coding region. A second promoterless GFP-donor plasmid contains a core target sequence upstream of a GFP reading frame (Supplementary Fig. S5B, C). Therefore, successful intermolecular recombination would result in the GFP donor being integrated downstream of the EF1α-HTLV promoter, resulting in dual-GFP and –mCherry positive cells (Fig. 4A). As such, we used GFP expression as detected by flow cytometry and fluorescence microscopy as an indicator of recombination efficiency. As expected, cotransfection of iCas9 and a nontarget sgRNA [sg(NT)] did not result in significant intermolecular molecular recombination with only mCherry expressing cells present (Fig. 4B–D). However, targeting with sgRNA pairs with 22 bp spacing [sg(G:H)] resulted in a significant increase in GFP-positive cells (Fig. 4B–D). Together, these results indicate that iCas9 is capable of both plasmid-based DNA deletion and integration in human cells.
FIG. 4.
iCas9 targets plasmid-to-plasmid recombination in human cells. (A) A dual-plasmid reporter for detection of intermolecular recombination. A promoterless GFP donor vector contains an iCas9 recognition site. A separate mCherry acceptor vector contains an EF1α-HTLV promoter with iCas9 target site and mCherry downstream. Recombination results in placement of GFP downstream of the promoter and mCherry-GFP dual-positive cells. (B) HEK293Ts were cotransfected with dual-reporter plasmids, iCas9 and sgRNAs. Scale bar = 200 μm. (C) Flow cytometry scatter plots of plasmid-to-plasmid recombination experiments. Untransfected HEK293Ts (gray, lower left, LL) were used to define gates for GFP+ and mCherry+ (dashed lines). HEK293Ts were cotransfected with reporter vectors, iCas9, and sg(NT) (red) or sg(G:H) (blue). Targeting resulted in GFP-mCherry dual-positive cells (upper right, UR). (D) Fold-increase of GFP-mCherry dual-positive cells for iCas9 transfections. Targeting of GFP-donor and mCherry-acceptor with sg(G:H) results in a 10.6 ± 0.5 fold-increase of dual-positive cells, results of recombination, at the target site compared to a control sgRNA sg(NT). ***p < 0.001, as determined by one-tailed Student's t-test.
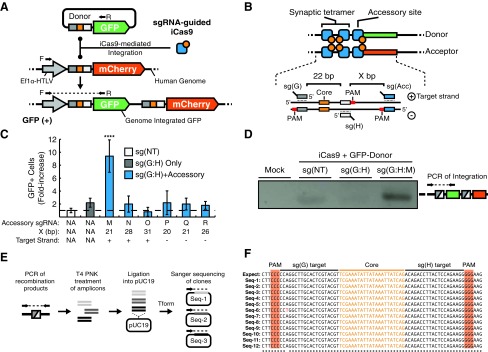
Given iCas9 targeted plasmid-to-plasmid recombination, we attempted to determine whether iCas9 can mediate plasmid-to-genome integration. To accomplish this proof-of-principle experiment, we adapted our plasmid-based assay to detect genome integration (Fig. 5A). Briefly, we used a retroviral vector (Supplementary Fig. S5D) to generate a stable HEK293T line in which the mCherry acceptor cassette (EF1α/HTLV hybrid promoter with a core target site upstream of a mCherry coding region) was integrated into the genome (Fig. 5A). Similar to the plasmid-based assay, successful genome integration of the GFP donor plasmid would result in in dual-GFP and –mCherry positive cells. As expected, when we cotransfected iCas9 and the GFP donor vector with a nontargeting sgRNA [sg(NT)], we did not observe recombination (Supplementary Fig. S6B). Surprisingly, when we cotransfected iCas9 and the GFP donor vector with a sgRNA pair [sg(G:H)] that previously allowed for integration in our plasmid-based assay, we did not observe recombination (Supplementary Fig. S6B).
FIG. 5.
Multiplex targeting of iCas9 enables genome integration in human cells. (A) A genome integrated mCherry acceptor cassette contains an elongation factor 1α–human T-cell leukemia virus hybrid promoter (EF1α-HTLV), and downstream iCas9 recognition site with an mCherry coding sequence. Integration of GFP into the genomic acceptor cassette results in GFP+ cells. (B) Design scheme for accessory targeting adjacent to the iCas9 core target site. Recombination between GFP-donor (green) and mCherry-acceptor (red) is coordinated by multiplex targeting of iCas9 binding. Accessory guide sites were targeted downstream of the iCas9 core site. We tested targeting of the + or − strand and varying distances (x bp). (C) Fold-increase of GFP+ over sg(NT) control. Targeting with iCas9 at the core site and downstream accessory 22 bp away resulted in 9.4 ± 2.5 fold-increase of GFP+ cells. Compared to sg(NT): ****p < 0.0001, by one-way ANOVA with Dunnett's multiple comparison test. (D) PCR detection of integration from isolated genomic DNA using primers flanking the recombination junction (inset by photo). “Mock” is a mock transfection of the mCherry-acceptor HEK293T cell line. iCas9 and GFP-donor were cotransfected with various guide combinations: G:H, 22 bp spacing without accessory guide; and G:H:M, 22 bp spacing with accessory targeting. (E) Subcloning strategy for isolation and characterization of recombination products. Recombination products were amplified via PCR from genomic DNA. Amplicons were phosphorylated by treatment with T4 polynucleotide kinase (T4 PNK) then ligated into pUC19. Ligation reactions were transformed (Tform) and plasmid clones were Sanger sequenced. (F) Alignments of Sanger sequenced recombination products against the expected recombination product (expect). Seq1–Seq12 indicate the expected recombination product free of indel mutations. Asterisks indicate nucleotides are consistent across all sequencing results.
Because iCas9 mediated plasmid-to-plasmid but not plasmid-to-genome recombination, we hypothesized that cooperative targeting might be necessary to enable iCas9-bound GFP-donor plasmids to interact with the genomic acceptor locus to facilitate integration. In this vein, previous work has shown that bacterial Tn3 resolvase uses cooperative binding at adjacent accessory sites to ensure efficient recombination of cointegrate products, where Tn3 resolvase coordinates substrate DNA bending, supercoiling, and 3-D positioning.18,37 We reasoned that multiplex sgRNAs targeting can recreate accessory site binding, which should allow for extra mTn3 domains to coordinate interaction between the GFP donor and the acceptor locus. To test this strategy, we designed a series of sgRNAs adjacent to the target core sites. These sgRNAs were targeted to either the “+” or “−” strand at varying base pair distances from the core target site (Fig. 5B and Supplementary Fig. S6A). We cotransfected these accessory sgRNAs with sg(G:H), GFP donor, and iCas9 into the mCherry-acceptor line. Of the accessory sgRNAs tested, we observed a significant increase in the number of GFP-positive cells when using an accessory sgRNA that targeted the “+” strand 21 bp from the core site [sg(M)] (Fig. 5C and Supplementary Fig. S6B). To confirm that integration occurred on the genomic level, we isolated genomic DNA and performed PCR with primers flanking the integration junction (Fig. 5A). A PCR product was only observed in conditions in which the optimized accessory sgRNA was employed [sg(G:H:M)] (Fig. 5D). To further confirm the identity of integration PCR products, we utilized a strategy of amplicon sub-cloning and sequencing of recombination products (Fig. 5E). Sequencing of PCR amplicons indicated the expected recombination product free of apparent indels (Fig. 5F). In sum, this data indicates that iCas9 is capable of not only plasmid-targeting but also plasmid-to-genome integration.
Discussion
Here we report the development of iCas9, a hyperactive mutant Tn3 resolvase fused to dCas9. We demonstrated iCas9's function in two eukaryotic hosts—yeast and human cells. We initially focused on developing an iCas9 capable of targeting eukaryotic genomic DNA. To accomplish this, we detected recombination using a yeast-based fluorescent reporter system. These experiments identified optimal symmetric spacings of 22 and 40 bp and asymmetric spacing of 31 bp. Interestingly, this is consistent with the Watson–Crick DNA structure being 10.5 bp per helix turn combined with the requirement for colocalization of mTn3 catalytic domains to the same helical face of the DNA molecule (Supplementary Fig. S3). Furthermore, our optimal sgRNA spacing of 22 bp is corroborated by zinc finger mTn3 fusions, which have an optimal spacing of 20–22 bp.22,27 In general, this phenomena is supported by FokI-dCas9 fusions that use 15 or 25 bp spacings, where sgRNA spacing match the requirement for FokI dimerization on opposite DNA helical faces.33 We demonstrated iCas9's function in human cells using a plasmid-based reporter and confirmed the functionality of the 22 bp sgRNA spacing. We also found 30 bp to be functional, which is consistent with previous reports using analogous recombinase-Cas9 designs.24 We reason these altered spacing stringencies may be due to the use of supercoiled plasmids as substrates, which may have different spacing preference than linear genomic DNA.
Targeting strategies developed here may allow iCas9 and analogous recombinase Cas9s to direct DNA integration. To test this method, we developed a plasmid-based intermolecular recombination assay. Using this assay, we found iCas9 is capable of targeting recombination between separate DNA molecules (Fig. 4). We expanded on these experiments to enable targeting of DNA integration into the human genome (Fig. 5). Recently Chaikind et al. engineered a recombinase Cas9 (recCas9) via fusion of dCas9 with Gin invertase.24 Similarly to recCas9, iCas9 was able to achieve plasmid and genomic DNA deletion. However, we wanted to explore the ability of iCas9 to facilitate genome integration, which has not previously been reported in other Cas9-based systems. To accomplish this, we adopted a scheme of accessory target site binding, wherein sgRNAs are targeted adjacent to the core sequence guides. Accessory binding sites for Tn3 resolvase have been implicated in regulating 3-D presentation of recombinase subunits, local DNA supercoiling and result in improved recombination efficiency.26,29 We designed a tiling of sgRNAs to recapitulate this with iCas9. Interestingly, we identified one multiplexing strategy that employed an accessory sgRNA [sg(M)] that utilized a spacing and orientation that approximates the 22 bp spacing observed between the Res1 core and adjacent accessory binding sites native to Tn3 transposon (Fig. 5).18,26,37 The ability of iCas9 to achieve genomic integration offers new applications not currently afforded by other Cas9-based recombinase systems.
Currently, the versatility of iCas9 to facilitate genomic integration is constrained by the requirement of accessory sgRNA binding (as there may not be “NGG” PAM sequences available with the optimal positioning) as well as the Tn3 Res1 core sequence to enable targeted recombination (which might not be located near the site of desired integration). In the future, the constraint of the need for and accessory sgRNA could be resolved through the engineering of iCas9 versions that employ highly versatile PAM variants of Cas9, such as xCas9.38,39 Specifically, xCas9 requires only an “NG” PAM sequence, which could drastically improve the versatility of iCas9 targeting. As it relates to the requirement for the core sequence, novel Tn3 catalytic domains that target new core sequences could be achieved in the future by recombinase directed evolution. For instance, substrate linked protein evolution enabled optimization of multiple serine recombinases catalytic domains for sites diverging from their native core sequences.40 More recently, a robust and highly sensitive strategy for recombinase directed evolution was developed via split gene reassembly of an antibiotic resistance cassette.41 Split gene reassembly has enabled structure reprogramming of Tn3 resolvase and generation of Gin invertase variants that recognize up to 3.77 × 107 unique DNA sequences.42,43 Likewise, this has expanded the targeting scope of β and Sin recombinase.44 Furthermore, directed evolution using these methods has enabled targeting of recombinases to disease relevant and human genome safe harbor sites.45 Moving forward, these techniques will be beneficial in characterizing and extending the targeting versatility of iCas9.
Admittedly, the current integration efficiency with iCas9 (i.e., 0.078 ± 0.021%) is low. It should be noted that this efficiency is comparable to previous reports with recCas9, which achieved 0.023 ± 0.013% with genomic DNA deletion.24 This indicates general improvement of genome targeting efficiency for recombinase-Cas9s as an important point of optimization necessary before practical application. We speculate that the low efficiencies might be due to genomic DNA that is constrained by epigenetic interactions and genomic localization.46,47 In particular, genomic spatial constraints may reduce Cas9 binding and prevent recombinase tetramerization. In future work, the optimization of the recombinase catalytic domain might serve as a strategy to increase recombination efficiency.16 For instance, ΦC31 integrase efficiency was improved by 5.5-fold via scanning mutagenesis.48 In regards to iCas9-site design, we employed heterodimeric recombinase sites (i.e., requiring targeting by two sgRNAs; Fig. 2C); however, it is possible that recombinase sites with symmetrical targeting via a single sgRNA may be more optimal. In addition, modification of dCas9 itself could potentially improve targeting efficiency as has been reported for base editing systems that employed a codon optimized dCas9. Another strategy to improve iCas9 genomic integration efficiency would be through the optimization of the interdomain linkers. Although we examined a subset of such linkers, a more extensive analysis of a broader set of interdomain linkers might be warranted given their effect on the efficiencies with other Cas9 heterologous fusions proteins.15,33 Finally, as is the case with other genome modification systems, iCas9 efficiency could be improved by increasing its cellular expression through improved delivery strategies (e.g., electroporation) or optimized promoter systems. Overall, comprehensive optimization of Tn3 catalytic domain, interdomain linker, dCas9 expression and site design will be beneficial to improve iCas9's targeting efficiency moving forward.
One of the potential advantages of iCas9 is that because the mTn3 catalytic domains of iCas9 preferentially operates via paired targeting by sgRNAs (Fig. 2C), it reduces the likelihood of unwanted off-target modifications. In this study, given iCas9's relatively low efficiency at targeting genomic loci, we speculated that off-target editing, if any, would be beyond the limits of detection. However, with improved targeting efficiency it is possible that iCas9 can induce off-target dimerization and recombination in a manner similar to that has been reported with hyperactive serine recombinases.43 If this becomes an issue with future iCas9 variants, directed evolution and rationally targeted mutagenesis of dimer interfaces may be employed to generate iCas9 variants that avoid spontaneous and off-target recombination. For example, to address off-target interactions, Gaj and colleagues used rational design and directed evolution of recombinases dimer interfaces to reduce off-target homodimerization by greater than 500-fold.36 Finally, as we continue to optimize iCas9 targeting efficiency, it will be important to have strategies to detect off-target recombination. We envision techniques such as split gene assembly which allow for sensitive detection of recombinase function will enable for accurate measurement of potential iCas9 off-target effects.41
One of the major benefits of iCas9 is that because of its fused recombinase functionality it does not directly rely on DSBs repair pathways such as NHEJ and HDR. In particular, HDR-based repair is dependent on cell division,9 which has limited CRISPR-targeted editing techniques in post-mitotic cells. In addition, it has been shown that DSB-dependent editing results in upregulation of p53, which reduces editing efficiency and induces apoptosis of edited cell populations.10,11 Furthermore, DSBs can lead to large, multiple kilobase deletions, insertions, and complex rearrangements.12,13 In fact, PCR and sequencing of iCas9 recombination products revealed that they were free of unwanted indel mutations (Figs. 2F and 5F). With further optimization of the versatility and efficiency, iCas9 derivatives may be helpful in editing cell types recalcitrant to DNA manipulations and avoiding the pitfalls of DSB-based editing strategies.
We envision that with continued development that iCas9 will be beneficial for synthetic biology in the construction and implementation of recombinase-based gene networks. Recombinase based gene networks are of increasing interest to synthetic biology.49 These systems can integrate multiple biological inputs and turn them into saved “DNA memory.” Recombinase based logic can be constructed in a way to imbue biological systems with Boolean logic functions or even 8-bit memory.49,50 These systems are capable of robust function.50,51 Development of new iCas9s via engineering of mTn3 dimer interfaces could generate orthogonal iCas9 variants, which would enable RNA-programmed recombinase-based gene networks. This may be useful for generating new and novel genetic circuits.
Conclusion
In summary, we present a framework for design and targeting of a Tn3 resolvase–dCas9 fusion (iCas9). We use iCas9 to target both DNA deletion and integration on genomic and episomal substrates. We identify sgRNA design parameters for DNA deletion and cooperatively targeted integration. With characterization of versatility and improved efficiency, it is feasible that recombinase-Cas9 systems like iCas9 be useful for a variety of biotechnology-related applications. Finally, this system will enable the development of new approaches to CRISPR-Cas9–based genome engineering and the creation of new synthetic gene networks.
Supplementary Material
Acknowledgments
We thank the DNASU sequencing core for DNA sequencing and the Mayo Clinic Flow Cytometry Core for FACS services. Plasmids will be made available to not-for-profit research upon request.
The authors confirm that all co-authors have reviewed and approved the manuscript. The authors affirm that the paper is original, not under consideration by any other journal and that the material has not been previously published. K.S.B. and X.W. devised the project. K.S.B designed and conducted experiments in yeast. K.S.B., N.B. and P.B. designed experiments in HEK293T cells. K.S.B., N.B., and Q.Z. conducted experiments in HEK293T cells. D.A.B. and X.W. managed the project. K.S.B., N.B., D.A.B., and X.W. wrote to manuscript.
Author Disclosure Statement
A patent application has been filed for material related to work in this article.
Funding Information
X.W. is supported by a grant from the U.S. Department of Health and Human Services National Institutes of Health (R01-GM106081 and R01-GM131405). D.A.B. is supported by the U.S. Department of Health and Human Services National Institutes of Health (grant R21-AG056706). N.B. is supported by the International Foundation for Ethical Research graduate fellowship.
Supplementary Material
References
- 1. Cong L, Ran FA, Cox D, et al. . Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. DOI: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mali P, Yang L, Esvelt KM, et al. . RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. DOI: 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zetsche B, Gootenberg Jonathan S, Abudayyeh Omar O, et al. . Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. DOI: 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotech. 2014;32:347–355. DOI: 10.1038/nbt.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brookhouser N, Raman S, Potts C, et al. . May I cut in? Gene editing approaches in human induced pluripotent stem cells. Cells. 2017;6 DOI: 10.3390/cells6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suzuki K, Tsunekawa Y, Hernandez-Benitez R, et al. . In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. DOI: 10.1038/nature20565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He X, Tan C, Wang F, et al. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucl Acids Res. 2016;44:e85 DOI: 10.1093/nar/gkw064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmid-Burgk JL, Höning K, Ebert TS, et al. CRISPaint allows modular base-specific gene tagging using a ligase-4-dependent mechanism. Nat Commun. 2016;7:12338 DOI: 10.1038/ncomms12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Orthwein A, Noordermeer SM, Wilson MD, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. DOI: 10.1038/nature16142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ihry RJ, Worringer KA, Salick MR, et al. p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat Med. 2018;24:939–946. DOI: 10.1038/s41591-018-0050-6 [DOI] [PubMed] [Google Scholar]
- 11. Haapaniemi E, Botla S, Persson J, et al. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24:927 DOI: 10.1038/s41591-018-0049-z [DOI] [PubMed] [Google Scholar]
- 12. Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotech. 2013;31:822–826. DOI: 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotech. 2018;36:765–771. DOI: 10.1038/nbt.4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. DOI: 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Komor AC, Zhao KT, Packer MS, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv. 2017;3:eaao4774 DOI: 10.1126/sciadv.aao4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gaj T, Sirk SJ, Barbas CF. Expanding the scope of site-specific recombinases for genetic and metabolic engineering. Biotechnol Bioeng. 2014;111:1–15. DOI: 10.1002/bit.25096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Standage-Beier K, Wang X. Genome reprogramming for synthetic biology. Front Chem Sci Eng. 2017;11:37–45. DOI: 10.1007/s11705-017-1618-2 [DOI] [Google Scholar]
- 18. Grindley NDF, Whiteson KL, Rice PA. Mechanisms of Site-Specific Recombination. Annu Rev Biochem. 2006;75:567–605. DOI: 10.1146/annurev.biochem.73.011303.073908 [DOI] [PubMed] [Google Scholar]
- 19. Brafman D, Willert K. Gene Transduction approaches in human embryonic stem cells. In: Methodological Advances in the Culture, Manipulation and Utilization of Embryonic Stem Cells for Basic and Practical Applications. (Atwood G, ed.) IntechOpen, 2011. DOI: 10.5772/14163 [DOI] [Google Scholar]
- 20. St-Pierre F, Cui L, Priest DG, et al. . One-step cloning and chromosomal integration of DNA. ACS Synth Biol. 2013;2:537–541. DOI: 10.1021/sb400021j [DOI] [PubMed] [Google Scholar]
- 21. Karpinski J, Hauber I, Chemnitz J, et al. . Directed evolution of a recombinase that excises the provirus of most HIV-1 primary isolates with high specificity. Nat Biotech. 2016;34:401–409. DOI: 10.1038/nbt.3467 [DOI] [PubMed] [Google Scholar]
- 22. Akopian A, He J, Boocock MR, et al. . Chimeric recombinases with designed DNA sequence recognition. PNAS. 2003;100:8688–8691. DOI: 10.1073/pnas.1533177100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mercer AC, Gaj T, Fuller RP, et al. . Chimeric TALE recombinases with programmable DNA sequence specificity. Nucl Acids Res. 2012:gks875 DOI: 10.1093/nar/gks875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaikind B, Bessen JL, Thompson DB, et al. . A programmable Cas9-serine recombinase fusion protein that operates on DNA sequences in mammalian cells. Nucl Acids Res. 2016;44:9758–9770. DOI: 10.1093/nar/gkw707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gordley RM, Gersbach CA, Barbas CF. Synthesis of programmable integrases. PNAS. 2009;106:5053–5058. DOI: 10.1073/pnas.0812502106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arnold PH, Blake DG, Grindley ND, et al. . Mutants of Tn3 resolvase which do not require accessory binding sites for recombination activity. EMBO J. 1999;18:1407–1414. DOI: 10.1093/emboj/18.5.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prorocic MM, Wenlong D, Olorunniji FJ, et al. . Zinc-finger recombinase activities in vitro. Nucl Acids Res. 2011;39:9316–9328. DOI: 10.1093/nar/gkr652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang W, Steitz TA. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. DOI: 10.1016/0092-8674(95)90307-0 [DOI] [PubMed] [Google Scholar]
- 29. Li W, Kamtekar S, Xiong Y, et al. . Structure of a synaptic γδ resolvase tetramer covalently linked to two cleaved DNAs. Science. 2005;309:1210–1215. DOI: 10.1126/science.1112064 [DOI] [PubMed] [Google Scholar]
- 30. DiCarlo JE, Norville JE, Mali P, et al. . Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucl Acids Res. 2013:gkt135 DOI: 10.1093/nar/gkt135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotech. 2009;27:465–471. DOI: 10.1038/nbt.1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotech. 2014;32:577–582. DOI: 10.1038/nbt.2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nishimasu H, Ran FA, Hsu PD, et al. . Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell. 2014;156:935–949. DOI: 10.1016/j.cell.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Standage-Beier K, Zhang Q, Wang X. Targeted large-scale deletion of bacterial genomes using CRISPR-nickases. ACS Synth Biol. 2015;4:1217–1225. DOI: 10.1021/acssynbio.5b00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gaj T, Sirk SJ, Tingle RD, et al. Enhancing the specificity of recombinase-mediated genome engineering through dimer interface redesign. J Am Chem Soc. 2014;136:5047–5056. DOI: 10.1021/ja4130059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nöllmann M, Byron O, Stark WM. Behavior of Tn3 resolvase in solution and its interaction with res. Biophys J. 2005;89:1920–1931. DOI: 10.1529/biophysj.104.058164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu JH, Miller SM, Geurts MH, et al. . Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. DOI: 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chatterjee P, Jakimo N, Jacobson JM. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci Adv. 2018;4:eaau0766 DOI: 10.1126/sciadv.aau0766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gordley RM, Smith JD, Gräslund T, et al. . Evolution of programmable zinc finger-recombinases with activity in human cells. J Mol Biol. 2007;367:802–813. DOI: 10.1016/j.jmb.2007.01.017 [DOI] [PubMed] [Google Scholar]
- 41. Gersbach CA, Gaj T, Gordley RM, et al. Directed evolution of recombinase specificity by split gene reassembly. Nucl Acids Res. 2010;38:4198–4206. DOI: 10.1093/nar/gkq125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gaj T, Mercer AC, Gersbach CA, et al. Structure-guided reprogramming of serine recombinase DNA sequence specificity. PNAS. 2011;108:498–503. DOI: 10.1073/pnas.1014214108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gaj T, Mercer AC, Sirk SJ, et al. A comprehensive approach to zinc-finger recombinase customization enables genomic targeting in human cells. Nucl Acids Res. 2013;41:3937–3946. DOI: 10.1093/nar/gkt071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sirk SJ, Gaj T, Jonsson A, et al. Expanding the zinc-finger recombinase repertoire: directed evolution and mutational analysis of serine recombinase specificity determinants. Nucl Acids Res. 2014;42:4755–4766. DOI: 10.1093/nar/gkt1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wallen MC, Gaj T, Iii CFB. Redesigning recombinase specificity for safe harbor sites in the human genome. PLOS ONE. 2015;10:e0139123 DOI: 10.1371/journal.pone.0139123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889 DOI: 10.1101/cshperspect.a003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brickner J. Genetic and epigenetic control of the spatial organization of the genome. Mol Biol Cell. 2017;28:364–369. DOI: 10.1091/mbc.E16-03-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liesner R, Zhang W, Noske N, et al. . Critical amino acid residues within the φC31 integrase DNA-binding domain affect recombination activities in mammalian cells. Hum Gene Ther. 2010;21:1104–1118. DOI: 10.1089/hum.2010.034 [DOI] [PubMed] [Google Scholar]
- 49. Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat Biotech. 2013;31:448–452. DOI: 10.1038/nbt.2510 [DOI] [PubMed] [Google Scholar]
- 50. Yang L, Nielsen AAK, Fernandez-Rodriguez J, et al. . Permanent genetic memory with >1-byte capacity. Nat Meth. 2014;11:1261–1266. DOI: 10.1038/nmeth.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weinberg BH, Pham NTH, Caraballo LD, et al. . Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nat Biotech. 2017;35:453–462. DOI: 10.1038/nbt.3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.