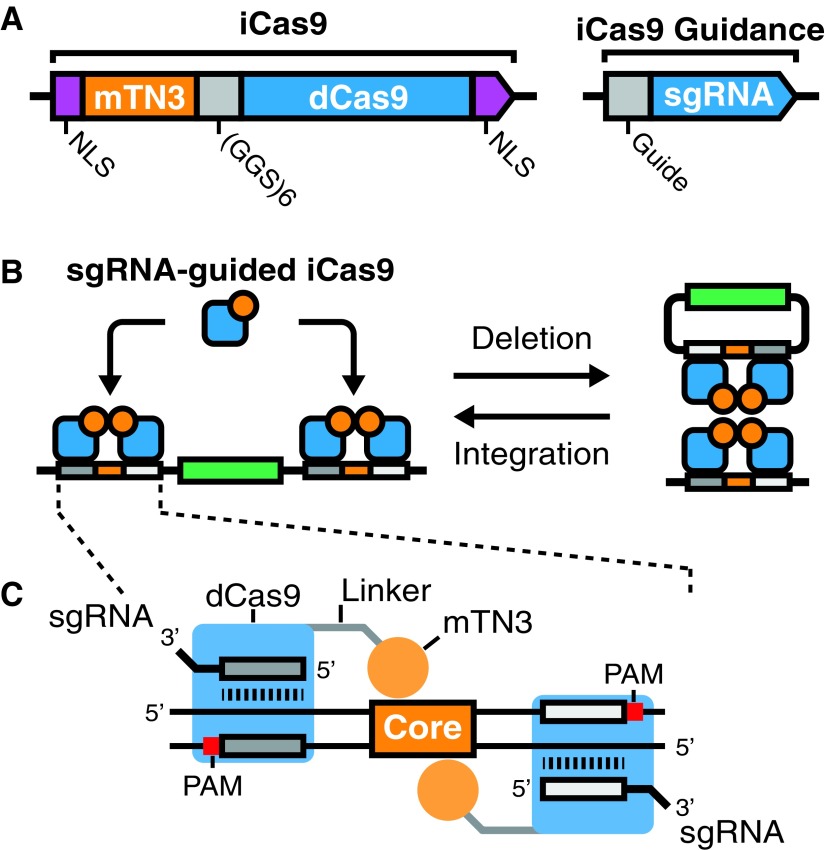
FIG. 1.
Design of integrase CRISPR-associated protein-9 nuclease (iCas9) and iCas9 target sites. (A) Architecture of the iCas9 fusion protein. A catalytically inactive Cas9 (dCas9, blue) is fused to the catalytic domain of a hyperactive mutant recombinase from transposon TN3 (mTN3, orange). dCas9 and mTN3 are separated by a flexible linker region (GGS × 6, gray). To promote nuclear entry, both N- and C-termini have SV40 nuclear localization signals sequences (NLS, purple). Given catalytic domains are fused to dCas9, iCas9 is guided via single guide RNAs (sgRNAs, blue). (B) mTN3 function is dependent on dimerization on target site sequences, followed by tetramerization. Tetramerization results in recombination, which can occur in two directions: deletion or integration. iCas9 can target either DNA deletion if target recognition sites are located on the same molecule (left), or alternatively iCas9 can target DNA integration if target sites are on separate DNAs (right). (C) The design of an iCas9 recognition site consists of two sgRNA targets (dark and light gray) flanking a TN3 Res1 core recognition sequence (core, orange). The two sgRNAs have a protospacer adjacent motif (PAM, red) distal orientation. mTN3–dCas9 fusions bind in positions around the core sequence allowing for mTN3 catalytic domain dimerization.