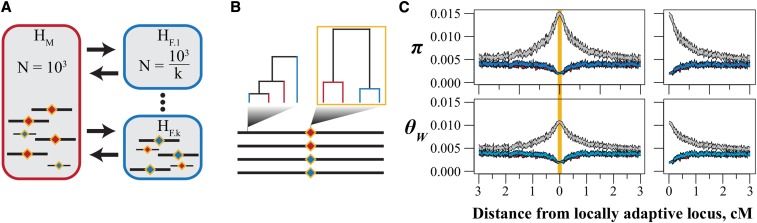
Figure 2.
The structure of simulations to test the effects of population structure on genetic variation in a model of local adaptation. (A) Habitat HM (red) consists of a single deme while habitat HF (blue) consists of k demes. We adjust the size of each deme in HF so that the total census size of each habitat is constant across simulations. Diamonds represent a single locus with locally adaptive alleles. (B) We assessed genetic diversity at the locally adaptive locus and in nonoverlapping windows. We used the pattern of reciprocal monophyly (outlined in gold) to classify RAD loci as divergent in the threespine stickleback population genomic dataset. [See Materials and Methods and Nelson and Cresko (2018)]. (C) Genetic diversity within (red and blue) and among (gray) allelic classes in a simulation where k = 1. Shown are 95% confidence intervals of nucleotide diversity (π) and Watterson’s θ after 10N generations of selection. Because patterns of variation are symmetrical about the locus under selection, we present “folded” curves [(C) right] throughout this work.