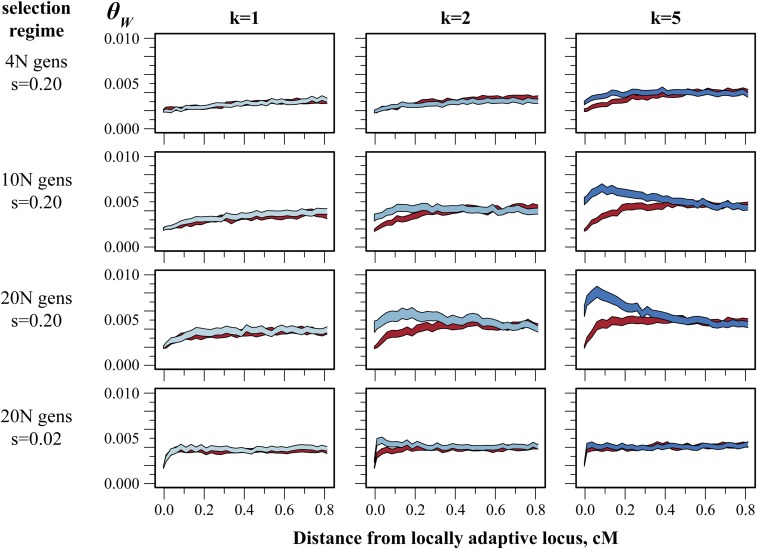
Figure 4.
Asymmetric population structure generates asymmetric patterns of linked variation on simulated chromosomes. Simulations were performed as described in Materials and Methods and Figure 2. Bands are 95% confidence intervals of Watterson’s θ at a given distance from a locally adaptive locus (red: θW on chromosomes carrying the HM-adaptive allele, blue: θW on chromosomes carrying the HF-adaptive allele). Columns show the effect of increasing population structure (k) in habitat HF. Rows show the effect of increasing the length of the selection phase of the simulations. Rows 1–3 show results under strong selection (s = 0.20). Row 4 shows results for simulations identical to row 3 but with moderate selection (s = 0.02). All simulations were performed using a migration rate, m, of one migrant per generation between habitat HM and each population of habitat HF. Total diversity (see Figure 2) is excluded to highlight variation within each allelic class.