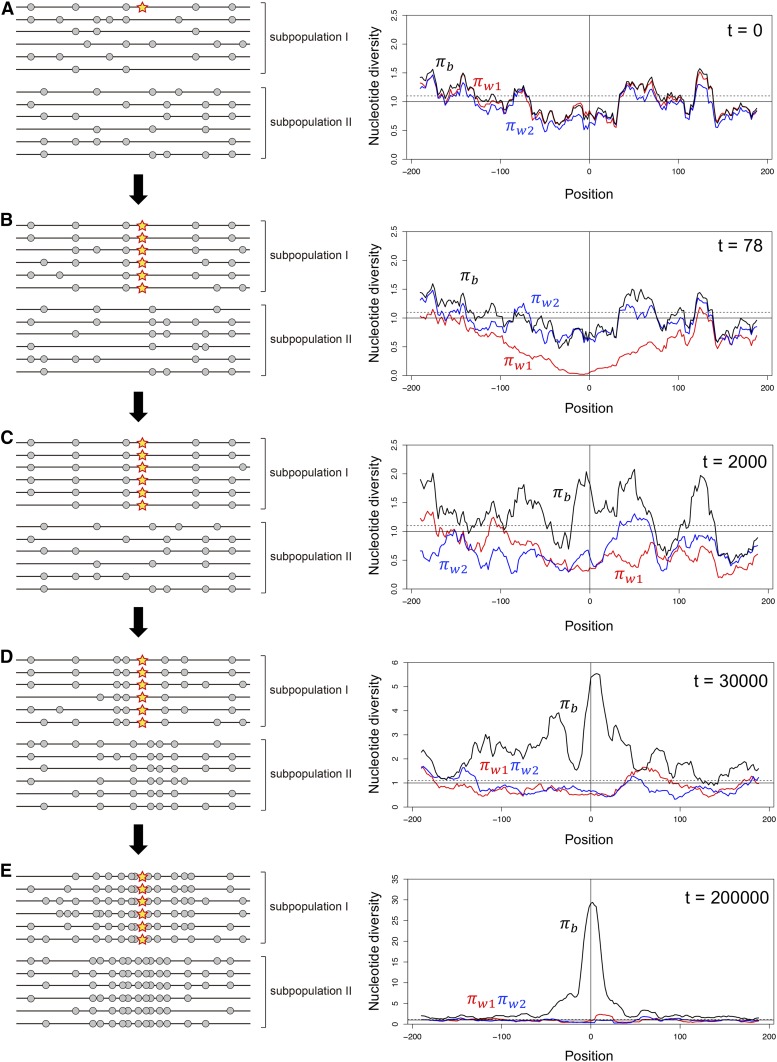
Figure 1.
Illustrating the evolution of a barrier locus in a simple two-population model with fairly high migration between them. (A) A locally adaptive de novo mutation arises in subpopulation I at position 0. A typical pattern of polymorphism is shown in left. The star is the locally adaptive mutation, and gray circles are neutral polymorphism in the surrounding region. The right panel shows the spacial distributions of nucleotide diversity obtained by a simulation. The simulation considers two subpopulations with population sizes are , between which symmetric migration is allowed at rate We assume selection intensity and . The entire simulated region is 400 kb if a population recombination rate of = 0.001 per site is assumed. See Appendix A for details about the simulation. The polymorphism levels within the two populations ( and ) are in red and blue, and divergence between the two populations is in black. , and can be considered as the averages of , and in a 20-kb window. The y-axis is adjusted such that under neutrality (the solid line) and the broken line exhibits , (i.e., = the genomic average). (B) The mutation quasi-fixes in subpopulation I, causing a drastic reduction in . (C) Migration shuffles polymorphisms in the two subpopulations, while selection works to maintain the quasi-fixation of the mutation. (D) The divergence gradually increases around the mutation, and (E) a clear peak of divergence arises. It should be noted that the star is fixed in the sample (left panes in B–E), but it does not necessarily mean that it is fixed in subpopulation I because there should be maladaptive immigrant alleles at a low frequency.