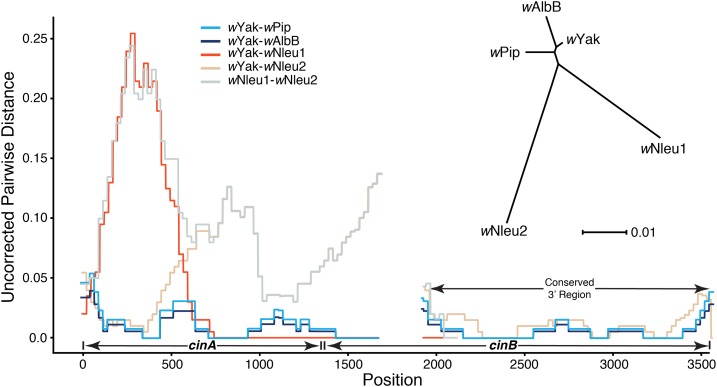
Figure 6.
The observed pairwise differences between cinA and cinB copies in wYak-clade Wolbachia and those in wAlbB and wPip, not including flanking regions, are shown. We also plot the pairwise differences between the two copies of these loci found in Nomada Wolbachia (denoted wNleu1 and wNleu2); we chose wNleu to represent the Nomada-clade Wolbachia because this strain contains the longest assembled contigs for both cinA-cinB copies. Homologous cinA-cinB copies have relatively low divergence (inset unrooted tree), with the highest divergence in the 5′ end of region. The most highly diverged region among any of the copies is the first ∼750 bp of wNleu1, with pairwise differences between wNleu1 and wYak reaching ∼25% in some windows, with similar divergence between wNleu1 and wNleu2 in this same region. Pairwise differences abruptly change 750 bp from the start of the cinA gene with the remaining ∼1150 bp of the assembled region having a single difference from the wYak sequence (only ∼1900 bp assembled into a single contig in the wNleu1 copy). This pattern suggests a recent transfer/recombination event from the same unknown donor of the wYak cinA-cinB copy. The gap at position 1705–1940 represents a tandem duplication present in wYak. The unrooted tree was generated using RAxML 8.2.9 (Stamatakis 2014), and the representative cinA-cinB gene copies were obtained using BLAST to search all Wolbachia genomes in the NCBI assemblies database (https://www.ncbi.nlm.nih.gov/assembly/?term=Wolbachia).