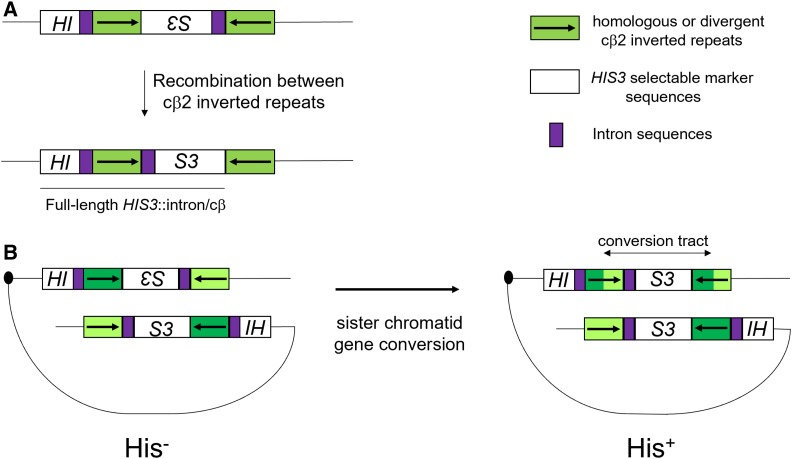
Figure 1.
Schematic of an intron-based recombination assay involving inverted repeat sequences that form a functional HIS3 reporter following homologous recombination [adapted from Nicholson et al. (2000)]. (A) Repeat sequences depicted by the green boxes are either identical in sequence (homologous substrate), or differ (divergent) by four SNPs (cβ2/cβ2-ns) or four 4-nt insertions (cβ2/cβ2-4L). The cβ2/cβ2-ns and cβ2/cβ2-4L substrates are predicted to form base–base and 4-nt loop mismatches in heteroduplex DNA, respectively. In this assay, spontaneous His+ colonies result from recombination between the 350-bp repeat sequences that reorient HIS3 (white boxes) and intron (purple boxes) sequences to yield a full-length HIS3::intron gene. (B) A model of how His+ recombinants arise in this system (Chen and Jinks-Robertson 1998). His+ recombinants are thought to result from gene conversion events between inverted repeat sequences present on sister chromatids that reorient HIS3 and intron sequences. In this example, divergent sequences are represented by different shades of green and the length of the gene conversion tract is shown. Homologous and divergent recombination rates for base–base and 4-nt loop substrates were calculated as described in the Materials and Methods, and the ratio of homologous to divergent recombination rates is presented as a measure of heteroduplex rejection efficiency.