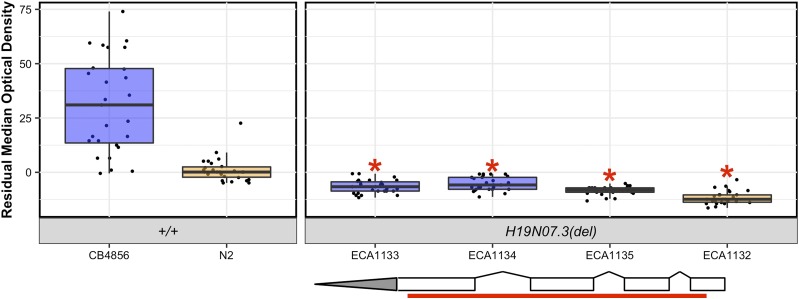
Figure 5.
Bleomycin responses of H19N07.3 deletion alleles are shown as Tukey box plots, with the strain name on the x-axis, split by genotype, and residual median optical density on the y-axis. Each point is a biological replicate. Strains are colored by their background genotype (orange indicates an N2 genetic background, and blue indicates a CB4856 genetic background). Two independent deletion alleles in each genetic background were created and tested. Red asterisks indicate a significant difference (P < 0.05, Tukey HSD) between a strain with a deletion and the parental strain that has the same genetic background. A depiction of the deletion allele is shown below the box plots. White rectangles indicate exons, and diagonal lines indicate introns. The 5′ and 3′ UTRs are shown by gray rectangles and triangles, respectively. The region of the gene that was deleted by CRISPR-Cas9 directed genome editing is shown as a red bar beneath the gene model.