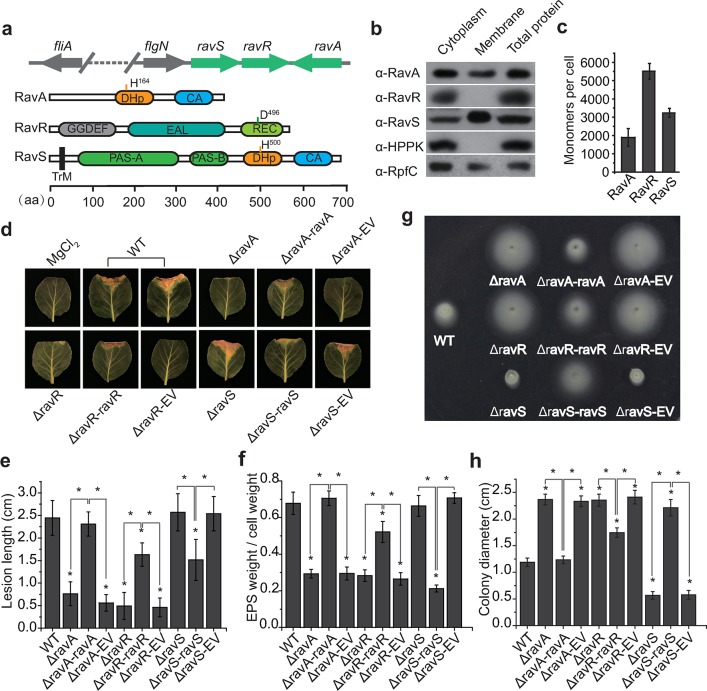
Fig 1. ravA, ravR and ravS differentially regulate bacterial virulence and swimming.
(a) Genomic organisation of ravARS genes and putative secondary structures of their protein products. Protein secondary structures were predicted by the SMART program. TrM: transmembrane domain; PAS: Per-ARNT-Sim domain; DHp: dimerisation and histidine phosphotransfer domain; CA: catalytic and ATP binding domain; REC: receiver domain. GGDEF and EAL domains are also shown. (b) Subcellular localisation of RavA, RavR and RavS. Western blotting was used to detect the proteins in different subcellular fractions. The known membrane-bound RpfC and cytoplasmic HPPK were detected as controls. (c) The ratio of RavA, RavR and RavS proteins in bacterial cells. Semi-quantitative western blotting was used to estimate the levels of these proteins (n = 3). (b) and (c), each experiment was repeated three times. (d–e) Mutation of ravA and ravR rather than ravS attenuated bacterial virulence significantly. (d) Bacterial virulence against host plant Brassica oleracea cv Zhonggan 11. Strains were inoculated onto plant leaves by scissor cutting. The lesion length was recorded 10 d after inoculation. The negative control was an inoculation of 10 mM MgCl2. EV: empty vector. (e) Quantification of the lesion length in (d). Average lengths and standard deviations are shown. Asterisk: significant difference, as tested by Student’s t-test (P ≤ 0.05, n = 30 inoculation sites). (f) Extracellular polysaccharide (EPS) production of bacterial strains. EPS production was measured as the dry weight of EPS vs. the dry weight of bacterial cells. Asterisk: significant difference, as tested by Student’s t-test (P ≤ 0.05, n = 3). (g–h) Swimming motility of bacterial strains. (g) Bacterial strains were inoculated in NYG plates containing 0.15% agar and grown at 28°C for 28 h. (h) Average diameters of the migration zones of (g). Asterisk: significant difference, as tested by Student’s t-test (P ≤ 0.05, n = 10).