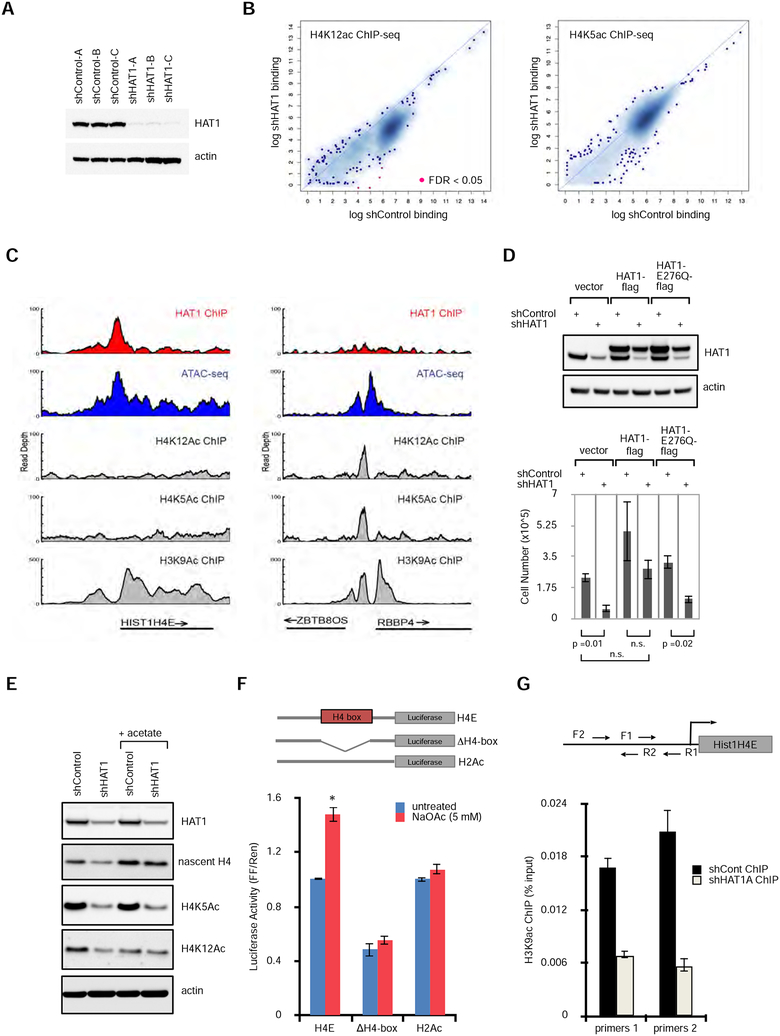
Figure 4: HAT1 promotes acetyl-transfer reactions to histone H3 at H4 promoters.
A. Stable cell lines expressing control shRNAs or three independent shRNAs to HAT1 were fractionated by SDS-PAGE, then immunoblotted.
B. ChIP-seq was performed with antibodies recognizing H4K12ac (left) or H4K5ac (right). Enriched loci were defined compared to input control sequencing. Loci that were significantly enriched or depleted are in red (FDR <0.05). See also Figure S4A.
C. Genome viewer tracks of read depth at the HIST1H4E (left) and the ZBTB8OS/RBBP4 (right) loci for HAT1 ChIP-seq, ATAC-seq, and histone modification ChIP-seq with antibodies that recognize acetylated lysines on H4K5, H4K12 and H3K9.
D. hTert-HME1 cells were treated with lentivirus expressing control or HAT1 shRNAs and also with lentiviruses expressing HAT1-flag, HAT1-E276Q-flag or vector control. (upper) Protein was fractionated by SDS-PAGE and immunoblotting was performed. (lower) Proliferation was measured by cell counting (mean +/− SD, n=3).
E. hTert-HME cells were infected with control or HAT1-targeted lentiviral shRNAs and maintained in culture for 6 days. Sodium acetate (5 mM) was added on day 4. Proteins were fractionated by SDS-PAGE and immunoblotted. See also Figure S4B.
F. Luciferase constructs were designed as indicated with 500 bp of the HIST1H4E promoter cloned upstream of Firefly luciferase. H4-box indicates that the 17 bp H4-box sequence was deleted. As a control 250 bp of the HIST1H2AC promoter was used. Constructs were co- transfected with a Renilla luciferase plasmid to 293T cells and luciferase activity (mean +/− SD) was measured 16 hours later. n=3 biological replicates. See also Figure S4C.
G. hTert-HME1 cells were infected with lentiviral control or HAT1-targeted shRNAs then six days later crosslinking and ChIP-qPCR was performed with antibodies to H3K9Ac and primer sets to the HIST1H4E promoter as indicated. Mean +/− SD, n = 3. The experiment has been independently replicated twice. See also Figure S4C.