Figure 5: HAT1 is required for locus-specific histone H3 acetylation.
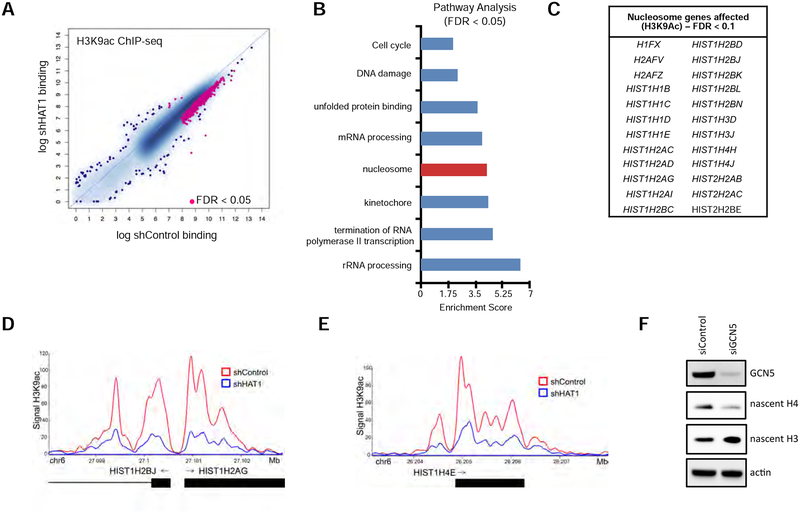
A. ChIP-seq was performed with antibodies to H3K9ac in six cell lines treated with either control or HAT1-targeted shRNAs (see Fig. 4A) and differential peak-calling was performed. Peaks differentially bound with FDR < 0.05 are indicated in pink. See also Figure S5A.
B. Pathway analysis of genes linked to altered H3 lysine 9 acetylation from Fig. 5A.
C. List of histone genes identified in the ‘nucleosome’ pathway from Fig. 5B to have significantly decreased H3 lysine 9 acetylation signal in the HAT1-depleted cells compared to control (FDR < 0.1).
D. Genome viewer track of H3 lysine 9 ChIP-seq signal at the HIST1H2BJ/HIST1H2AG locus in shControl and shHAT1-treated cells.
E. Genome viewer track of H3 lysine 9 ChIP-seq signal at the HIST1H4E locus in shControl and shHAT1-treated cells.
F. hTert-HME1 cells were transfected with control or KAT2A-targeted siRNAs and three days later proteins were harvested by detergent extraction and immunoblotting was performed. See also Figure S5B, C.