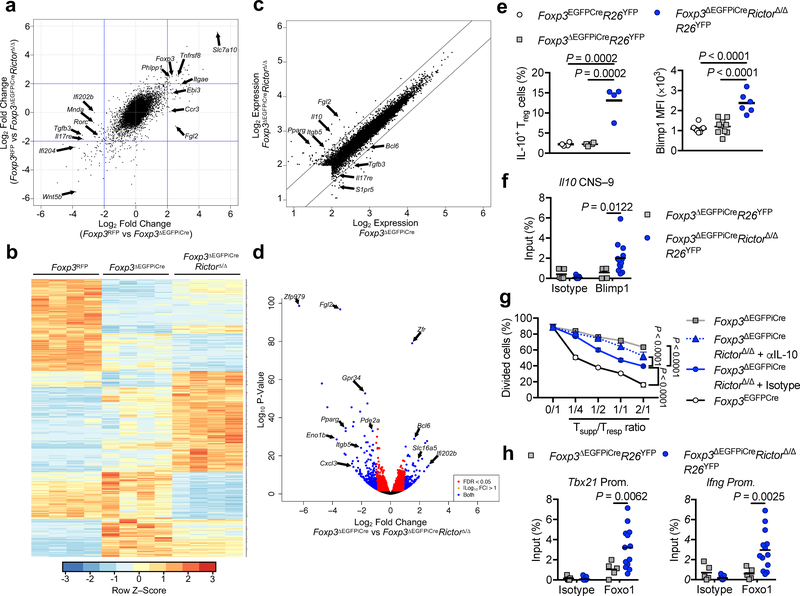
Figure 4. mTORC2-dependent and -independent gene expression profiles in ΔTreg cells.
Gene expression profiles of Foxp3RFP (WT), Foxp3ΔEGFPiCre and Foxp3ΔEGFPiCreRictorΔ/Δ Treg cells (n=4 per group) isolated from spleen of Foxp3RFP/ΔEGFPiCreR26YFP and Foxp3ΔEGFPiCre/+RictorΔ/ΔR26YFP female mice. (a), Gene expression profiles represented as Fold change (Foxp3RFP vs Foxp3ΔEGFPiCre) versus Fold change (Foxp3RFP vs Foxp3ΔEGFPiCreRictorΔ/Δ). (b), Heatmap representation, (c), Log2 expression and (d) volcano plot of differential gene expression in Foxp3ΔEGFPiCre versus Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells. FDR, false discovery rate; log2FC, log2(fold change). (e), Flow cytometric analysis of IL-10 and Blimp1 expression in Treg cells of Foxp3EGFPCre (n=4 and n=8 respectively) and ΔTreg cells of Foxp3ΔEGFPiCre (n=4 and n=10 respectively) and Foxp3ΔEGFPiCreRictorΔ/Δ (n=4 and n=6 respectively) mice. Results are expressed as scatter plot and mean and represent 1 of 3 independent experiments. (f), Chromatin immunoprecipitation (ChIP) assay for the binding of Blimp1 and control isotype to the Il10 conserved non-coding sequence located 9kb in proximal of il10 promoter (il10 CNS–9) in Foxp3ΔEGFPiCre (n=5) and Foxp3ΔEGFPiCreRictorΔ/Δ (n=13) ΔTreg cells. Results are expressed as scatter plot and mean and represent a pool of 2 independent experiments. (g), In vitro suppression of Tresp cells by Foxp3ΔEGFPiCreRictorΔ/Δ ΔTreg cells carried out in the presence of either an isotype control or anti-IL-10 mAb (n=3 per group). Results are expressed as mean ± SEM and represent 1 of 2 independent experiments. (h) ChIP assays for the binding of Foxo1 and control isotype to Tbx21 and Ifng promoters in Foxp3ΔEGFPiCre (n=5) and Foxp3ΔEGFPiCreRictorΔ/Δ (n=13) ΔTreg cells. Results are expressed as scatter plot and mean and represent a pool of 2 independent experiments. Statistical significance was determined one-way ANOVA (e) or two-way ANOVA (f-h) with Tukey’s or Sidak’s multiple comparisons (P values as indicated).