Figure 6. The +41 kb Irf8 enhancer is required for cDC1 specification and development.
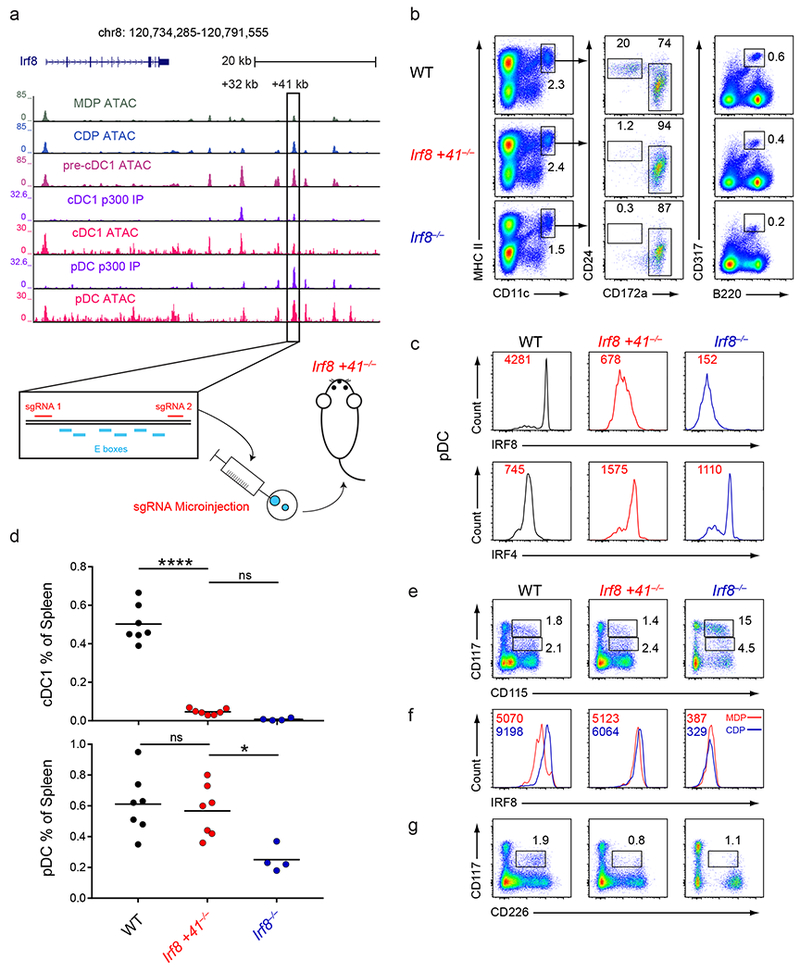
a, Normalized sequencing tracks of ChIP-seq with anti-p300 antibody or of ATAC-seq in the indicated populations. Boxed is the +41 kb Irf8 enhancer. Shown below is the +41 kb Irf8 enhancer with the E box motifs and sgRNA target sequences indicated. ATAC-seq data are representative of three independent experiments with similar results (n = 3 biological replicates for MDP ATAC, CDP ATAC, and pre-cDC1 ATAC) and ChIP-seq data are pooled from two independent experiments and the Immunological Genome Project Open Chromatin Regions (n = 1 biological replicate for cDC1 p300 IP, pDC p300 IP, cDC1 ATAC, and pDC ATAC). b, Flow cytometry of live splenocytes from mice of the indicated genotypes was used to identify DC subsets. Numbers indicate the percent of cells in the indicated gates. Data are representative of five independent experiments with similar results (n = 6 mice for WT, n = 7 mice for Irf8 +41−/−, and n = 4 mice for Irf8−/−). c, Intracellular staining for IRF8 and IRF4 in splenic pDCs of mice of the indicated genotypes. Numbers indicate the MFI of IRF8 or IRF4 protein levels in pDCs. Data are representative of four independent experiments with similar results (n = 5 mice for WT, n = 5 mice for Irf8 +41−/−, and n = 3 mice for Irf8−/−). d, Statistical analysis of the frequency of cDC1s and pDCs in spleens of mice of the indicated genotypes. Small horizontal lines indicate the mean. Data are pooled from five independent experiments (n = 6 mice for WT, n = 7 mice for Irf8 +41−/−, and n = 4 mice for Irf8−/−). e, Flow cytometry of Lin−CD135+ BM from mice of the indicated genotypes was used to identify MDPs and CDPs. Numbers indicate the percent of cells in the indicated gates. Data are representative of three independent experiments with similar results (n = 3 mice for WT, n = 3 mice for Irf8 +41−/−, and n = 3 mice for Irf8−/−). f, Intracellular staining for IRF8 within MDPs and CDPs from mice of the indicated genotypes. Red lines indicate the IRF8 protein level in MDPs and blue lines indicate the IRF8 protein level in CDPs. Numbers in red indicate the MFI of IRF8 protein levels in the MDP and numbers in blue indicate the MFI of IRF8 protein levels in the CDP. Data are representative of three independent experiments with similar results (n = 3 mice for WT, n = 3 mice for Irf8 +41−/−, and n = 3 mice for Irf8−/−). g, Flow cytometry of Lin−CD135+ BM from mice of the indicated genotypes was used to identify pre-cDC1s. Numbers indicate the percent of cells in the indicated gates. Data are representative of three independent experiments with similar results (n = 3 mice for WT, n = 3 mice for Irf8 +41−/−, and n = 3 mice for Irf8−/−). ns, not significant (P>0.05); *P<0.05; and ****P<0.0001, ordinary one-way ANOVA (d).
