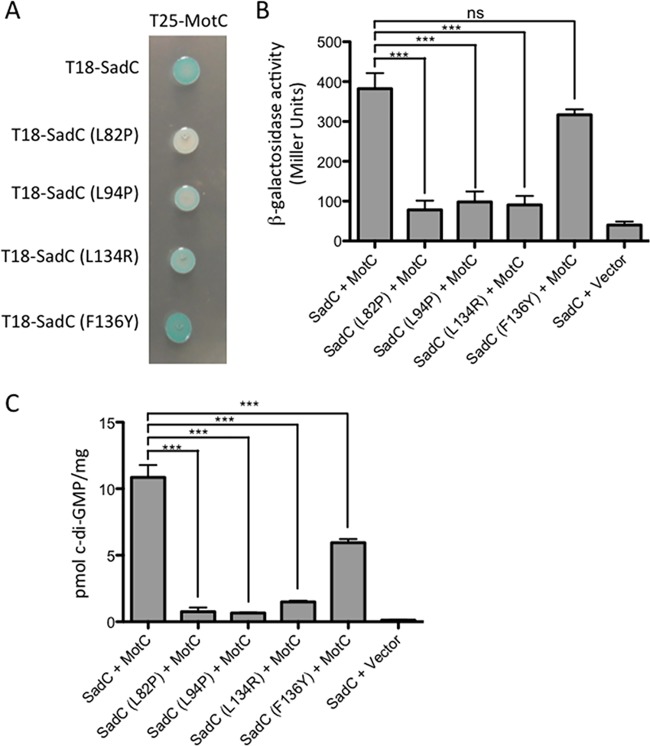
FIG 4.
Point mutations in the transmembrane domain of SadC disrupt both its interaction with MotC and c-di-GMP production. (A) The wild-type sadC gene and point mutant variants were cloned into the vector pUT18C. The motC gene was cloned into the vector pKT25. Plasmids were cotransformed into E. coli BTH101 cells. The transformants were spotted (2 μl) onto LB agar containing Cb, Kan, IPTG, and X-Gal. Plates were incubated at 30°C for 30 h. Cleavage of X-Gal (blue) indicates a positive protein-protein interaction. (B) Bacterial two-hybrid interactions were quantified by measuring the β-galactosidase activity of transformants grown at 30°C overnight in LB broth supplemented with Cb and Kan. “Vector” indicates an empty pKT25 vector. The data represent results from two experiments, each performed in triplicate. Values are means ± SEM. Significance was determined by analysis of variance and Dunnett’s posttest comparison for difference from the wild-type interaction (T18-SadC plus T25-MotC). ***, P < 0.001; ns, not significant. (C) Quantification of cellular c-di-GMP levels by LC-MS from B2H assays. The x axis displays the two fusion proteins (listed, in order, as pUT18C and pKT25) cotransformed into BTH101 cells. After being cotransformed with 2 fusion plasmids, cells were spotted onto LB agar containing Cb, Kan, and IPTG (with no X-Gal). Plates were incubated at 30°C for 40 h, and then cells were scraped off plates and nucleotides were extracted. Data are expressed as picomoles of c-di-GMP per milligram (dry weight) of cells from which nucleotides were extracted. Data represent results from three experiments, each performed in triplicate. Values are means ± SEM. Significance was determined by analysis of variance and Tukey’s post hoc test comparison for differences between the strains indicated. ***, P < 0.001.