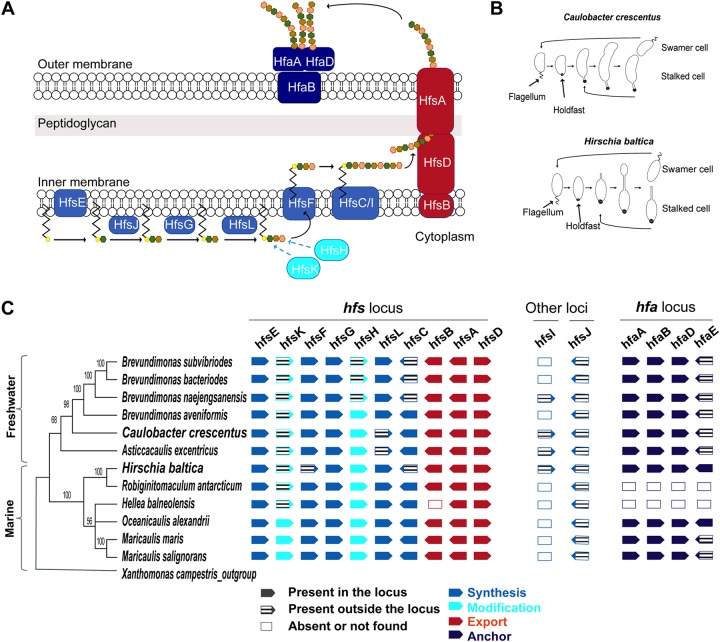
FIG 1.
Organization of the holdfast gene cluster in H. baltica. (A) Schematic of holdfast synthesis, modification, secretion, and anchor machineries. Holdfast polysaccharide synthesis is initiated by the glycosyltransferase HfsE, which transfers activated sugar precursors in the cytoplasm to a lipid carrier. Three glycosyltransferases, HfsJ, HfsG, and HfsL, add different sugars to the growing polysaccharide. The acetyltransferase HfsK and the deacetylase HfsH modify one or more sugar residues, and then a flippase, HfsF, transports the lipid carrier into the periplasm. Repeat units are polymerized by polymerases HfsC and HfsI. The polysaccharide is exported outside the cell through the HfsA-HfsB-HfsD complex. The exported polysaccharide is then anchored to the cell body by the secreted proteins HfaA, HfaB, and HfaD. The different colored hexagons represent different sugars. (B) Diagrams of C. crescentus and H. baltica dimorphic cell cycles. A motile swarmer cell differentiates into a stalked cell by shedding its flagellum and synthesizing a holdfast at the same cell pole. C. crescentus stalked cells divide asymmetrically to produce a motile swarmer and a stalked cell (top), and H. baltica reproduces by budding a motile swarmer off the stalk (bottom). (C) Maximum-likelihood phylogeny inferred from 16S rRNA sequences of selected freshwater and marine members of Caulobacterales. The node values represent clade frequencies of 1,000 bootstraps. The genes were identified using reciprocal best-hit analysis on fully sequenced Caulobacterales genomes. Solid gene symbols represent genes within the hfs or hfa loci, while hatched symbols indicate the genes translocated from these loci to a different location in the genome. Empty boxes indicate absent or missing genes.