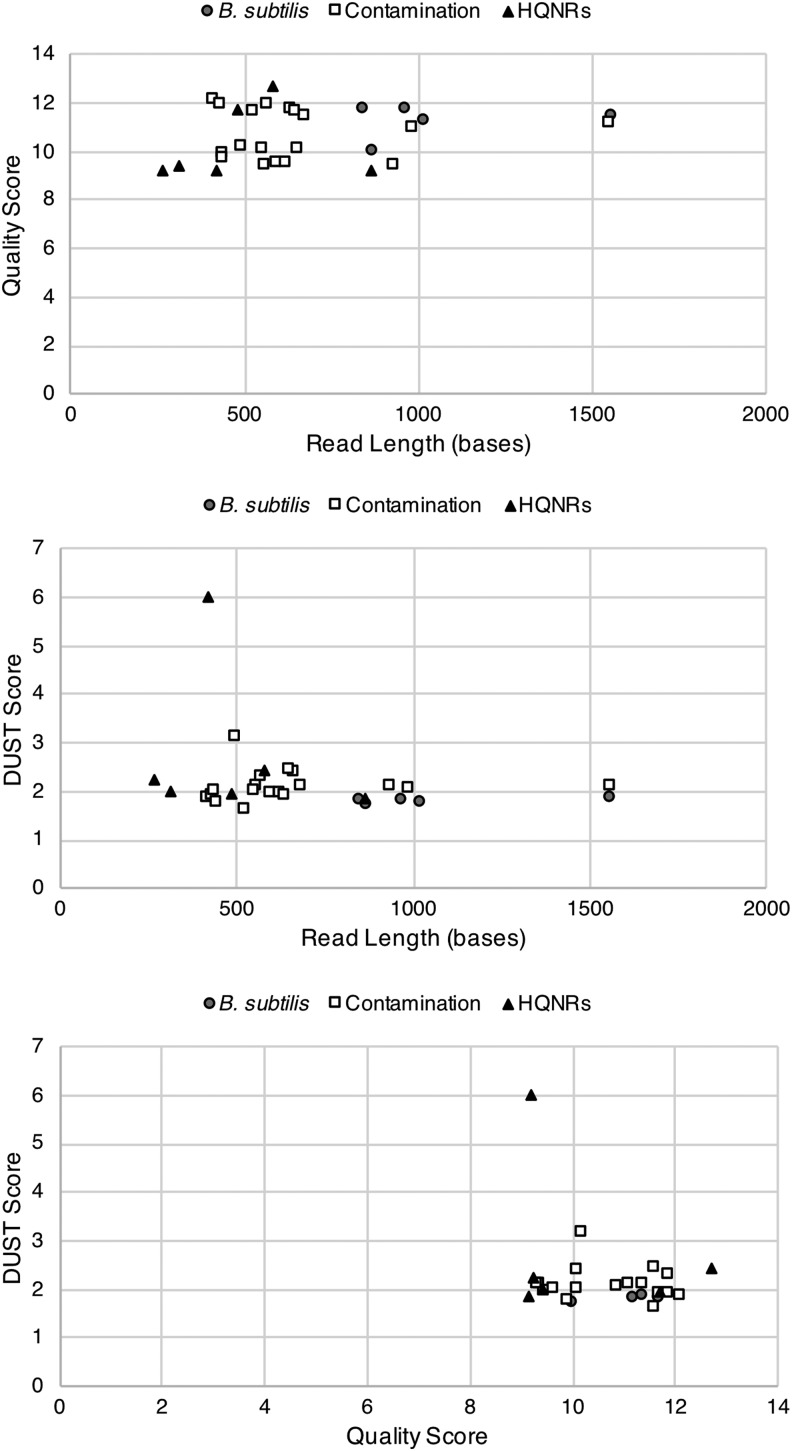
FIG. 4.
Cross-plots of target reads identified by CarrierSeq displaying read length, quality score, and DUST score. There does not appear to be any separation of HQNRs and B. subtilis or contamination reads. We are presently only able to identify HQNRs through k-mer matching or via the NCBI blastn algorithm.