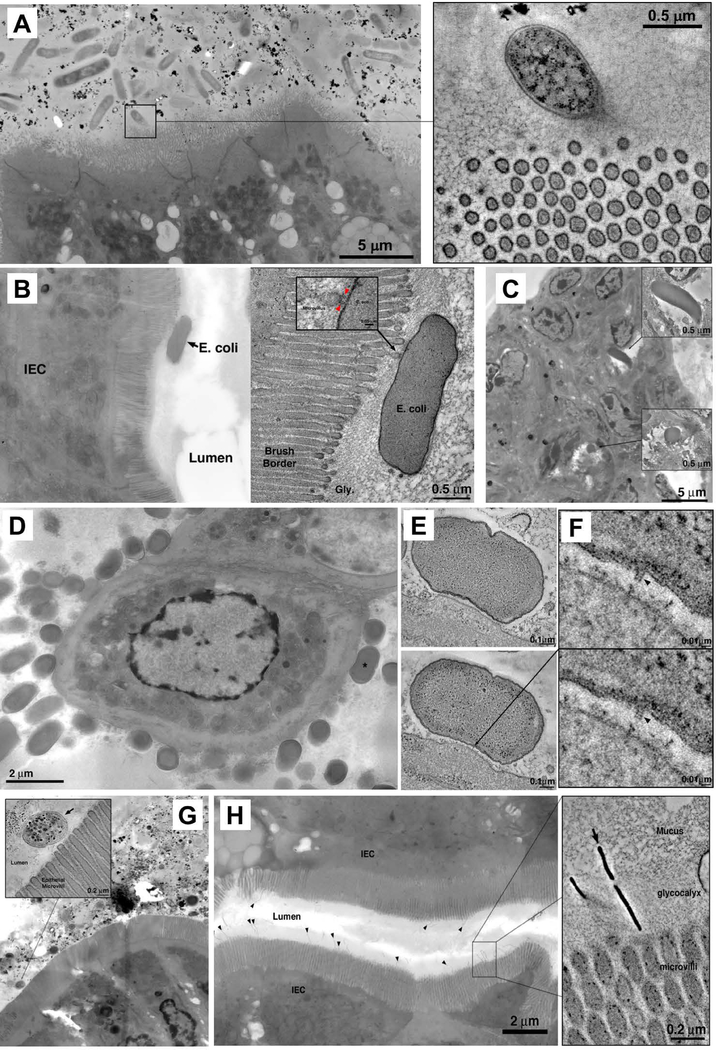
Fig 4. Electron tomography of host-microbe interactions in the intestine.
(A) Microbiota in the terminal ileum of SFB-negative Jackson C57BL/6 mice posess limited interactions with IECs and lack MATE. Inset: rare interaction between a bacterial cell and IEC microvilli. (B,C) Interactions of adherent-invasive Escherichia coli (AIEC) strains LF82 (B) and NRG857c (C) with IECs in terminal ileum. (A, right) Tomogram showing interaction with microvilli. (C) Internalized AIEC cells. (D) Citrobacter rodentium, overview showing numerous bacterial cells associating with IEC surface in the colon. (E) Tomograms of a Citrobacter pedestal. (F) Tomographic details of a presumptive T3SS needle connecting the microbe to IEC (arrowheads). (G) Mixture of 20 human Th17 cell-inducing strains. Inset; tomogram showing bacterial cells in the lumen, close to microvilli, but not making contact with IECs. (H) Bifidobacterium adolescentis (arrowheads) in terminal ileum. Inset: tomogram of B. adolescentis showing penetration of the glycocalyx and interaction with the IEC microvilli.