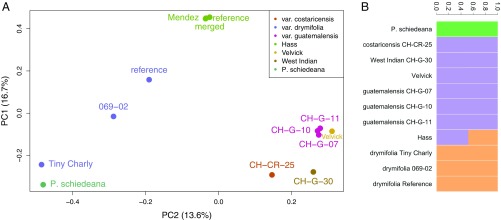
Fig. 1.
Population genomic structure of avocado. (A) Principal component analysis (PCA) of genome-wide SNPs reveals population groupings among races and varieties. The Guatemalan and West Indian/Costa Rican accessions are closely related, while the Mexican (P. americana var. drymifolia) specimens are more diverse, with the unusual individual Tiny Charly drawn toward the outgroup species P. schiedeana by PC2. Hass and its sport Mendez are tightly clustered and intermediate between Mexican and Guatemalan and West Indian/Costa Rican on PC2. (B) NGSAdmix analysis reveals similar population structure at K = 3. The P. schiedeana outgroup is distinct, and the Hass reference genome is revealed to be admixed between Guatemalan–West Indian and Mexican source populations, the Mexican source clearly contributing greater than 50%.