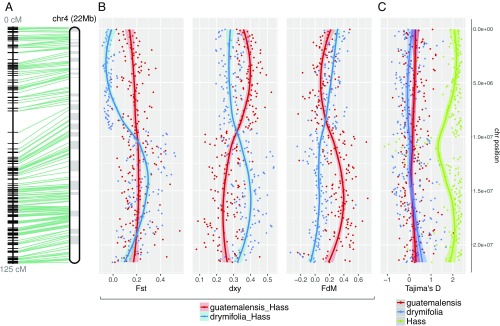
Fig. 2.
SNP diversity analysis reveals the hybrid genomic background of Hass avocado. (A) Twenty-two megabases (Mb) of anchored DNA on chromosome 4 exemplify the hybrid nature of Hass, in which genomic introgression from the Guatemalan avocado race (var. guatemalensis) occurred into a Mexican (var. drymifolia) genetic background. (B) While in the top chromosome arm the blue trend line shows a low differentiation index (FST) between Hass and the Mexican subpopulation as well as a high introgression signal from var. drymifolia into Hass, these signals should not be misinterpreted as introgression events, since the absolute genetic divergence between both sets of accessions does not vary along the chromosome. The lower arm of the chromosome, however, has inverted trends, where our estimators describe an elevated introgression signal from var. guatemalensis into Hass, as confirmed by the decay in (red trend line), and higher FST between Hass and Mexican accessions. (C) No evidence for selective sweeps or domestication signatures were identified; Mexican and Guatemalan subpopulations displayed neutral D values while Hass maintained extreme D values at the theoretical upper limit of the estimator (∼2). Such positive values reflect a “bottlenecked” origin with clonal expansion after the very recent foundation of the cultivar only a few decades ago. Each dot in the plots corresponds to statistics for SNP data in nonoverlapping 100-kb windows (confidence interval of 0.90 for graphical smoothed conditional means). Apparent centromeric regions are located at around 10 Mb, where FST, and intersect and Tajima’s D for Hass decreases.