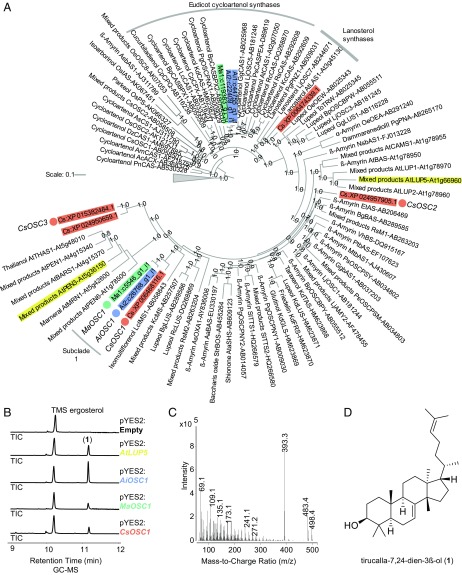
Fig. 2.
Identification and characterization of OSCs from limonoid-producing species. (A) Phylogenetic tree of candidate OSCs from A. indica (blue), M. azedarach (green), and C. sinensis (orange). Functionally characterized OSCs from other plant species (20) are included, with the 2 previously characterized tirucalla-7,24-dien-3β-ol synthases from A. thaliana (AtLUP5 and AtPEN3) highlighted (yellow). Human and prokaryotic OSC sequences used as an outgroup are represented by the gray triangle. Candidate OSCs chosen for further analysis are indicated (circles). The phylogenetic tree was constructed by FastTree V2.1.7 (68) and formatted using iTOL (69). Local support values from FastTree Shimodaira-Hasegawa (SH) test (between 0.6 and 1.0) are indicated at nodes, and scale bar depicts estimated number of amino acid substitutions per site. (B) GC-MS total ion chromatograms of derivatized extracts from yeast strains expressing candidate OSCs. Traces for the empty vector (pYES2) and strains expressing the candidates AiOSC1 (blue), MaOSC1 (green), CsOSC1 (orange), and the previously characterized AtLUP5 (yellow) are shown. (C) GC-MS mass spectra of TMS-tirucalla-7,24-dien-3β-ol (1). (D) Confirmation of the structure of the cyclization product generated by AiOSC1 as tirucalla-7,24-dien-3β-ol (1) by NMR (SI Appendix, Table S3). Traces, mass spectra, and product structure for CsOSC2 and CsOSC3 are given (SI Appendix, Figs. S2 and S3).