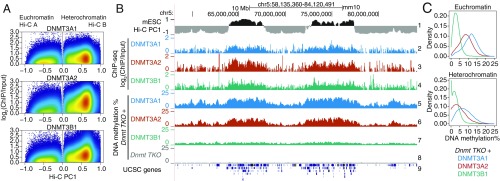
Fig. 6.
Contributions of DNMT3 enzymes to DNA methylation in euchromatin and heterochromatin. (A) Genome-wide distribution of the de novo DNA methyltransferases DNMT3A1, DNMT3A2, and DNMT3B1 within euchromatin and heterochromatin compartments in WT mESCs. DNMT ChIP-seq enrichment (log2 fold change) was calculated at 1-kb resolution. Data from refs. 51 and 52. (B) Genome tracks showing the distribution of DNMT3A1, DNMT3A2, and DNMT3B1 (tracks 2–4) within Hi-C defined compartments (track 1), and the contribution to DNA methylation of the de novo DNMT proteins after reconstitution in a Dnmt TKO background in mESCs (tracks 5–7). Dnmt TKO methylation is shown for reference (track 8). (C) Density distribution of average DNA methylation values within 10-kb windows across the euchromatin and heterochromatin compartments, in Dnmt TKO mESCs reconstituted with DNMT3A1, DNMT3A2, and DNMT3B1.