Fig. 2.
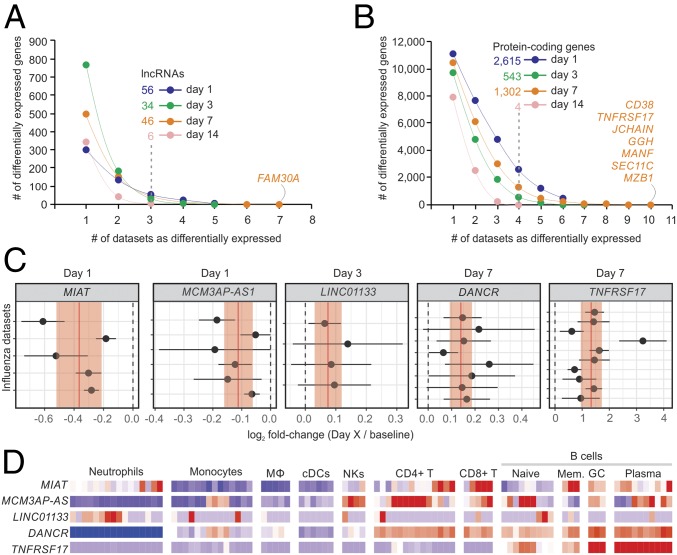
Transcriptome analysis of cohorts immunized with inactivated IV. Cumulative sum of differentially expressed lncRNAs (A) and protein-coding genes (B) (y-axis) in one or more datasets (x-axis). Transcripts were considered differentially expressed if limma P values were lower than 0.05 in at least three cohorts (for lncRNAs) or four cohorts (for protein-coding genes). (C) Forest plots of representative lncRNAs and TNFRSF17 with reiterated differential expression. Log2 fold changes with their corresponding 95% confidence interval (x-axis) are plotted for each cohort (y-axis). Red vertical lines and shaded regions represent log2 fold-change summaries and their 95% confidence intervals, respectively. (D) Heatmap depicting gene expression of human immune cells from ref. 28. Logs (FPKM) of genes are scaled around zero. Columns represent samples; rows represent genes.