Abstract
The subgenus Sophophora of Drosophila, which includes D. melanogaster, is an important model for the study of molecular evolution, comparative genomics, and evolutionary developmental biology. Numerous phylogenetic studies have examined species relationships in the well-known melanogaster, obscura, willistoni, and saltans species groups, as well as the relationships among these clades. In contrast, other species groups of Sophophora have been relatively neglected and have not been subjected to molecular phylogenetic analysis. Here, we focus on the endemic African Drosophila fima and dentissima lineages. We find that both these clades fall within the broadly defined melanogaster species group, but are otherwise distantly related to each other. The new phylogeny supports pervasive divergent and convergent evolution of male-specific grasping structures (sex combs). We discuss the implications of these results for defining the boundaries of the melanogaster species group, and weigh the relative merits of “splitting” and “lumping” approaches to the taxonomy of this key model system.
Keywords: Drosophila, Sophophora, melanogaster, fima, dentissima, sex combs
Graphical Abstract
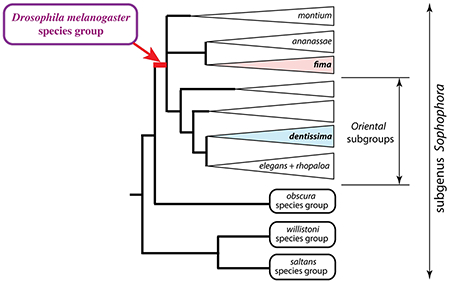
1. Introduction
Drosophila taxonomy is all the more complicated for the amount of attention it has attracted over the last century (O’Grady and DeSalle, 2018). The family Drosophilidae Rondani contains over 70 genera and well over 4000 described species, >1600 of which are assigned to the genus Drosophila Fallen (bioinfo.museum.hokudai.ac.jp/db/). It is abundantly clear that the “genus Drosophila” is in fact paraphyletic with respect to many of the smaller drosophilid genera. Although establishing a sounder classification based on monophyletic genera and subgenera could bring some order to this chaos, attempts at revision have been complicated by the sheer scale of this problem, by limited taxon sampling in molecular phylogenetic studies, and by the difficulty of resolving basal phylogenetic relationships (O’Grady, 2010; O’Grady and DeSalle, 2018; O’Grady and Markow, 2009; Remsen and O’Grady, 2002; van der Linde et al., 2010; Yassin, 2013).
The subgenus Sophophora of Drosophila, which was established by Alfred Sturtevant (Sturtevant, 1939) and includes the model species D. melanogaster, has long been an island of stability among the general turmoil of Drosophila systematics (O’Grady and Kidwell, 2002). Recently, however, Sophophora was found to be paraphyletic with respect to the genus Lordiphosa (Gao et al., 2011), which currently includes 69 described species (Fartyal et al., 2017; Katoh et al., 2018). The best known species groups of Sophophora are the melanogaster, obscura, willistoni, and saltans groups (Gao et al., 2003; Gleason et al., 1998; Lakovaara and Saura, 1982; Lemeunier et al., 1986; O’Grady, 1999; O’Grady et al., 1998; Robe et al., 2010; Rodriguez-Trelles et al., 1999). Molecular phylogenetic analyses have shown that the Neotropical willistoni and saltans species groups are more closely related to Lordiphosa than to the predominantly Old-World melanogaster and obscura species groups (Gao et al., 2011; Hu and Toda, 2000; Katoh et al., 2000).
Within Sophophora, the melanogaster species group has received by far the most attention. Its inclusion of the genetic model system D. melanogaster has stimulated wide-ranging work on the molecular evolution, comparative genomics, and evolutionary developmental biology of this lineage (Atallah et al., 2012; Barmina and Kopp, 2007; Chen et al., 2010; Chen et al., 2014; Jeong et al., 2006; Kursel and Malik, 2017; Levine et al., 2012; Levine et al., 2016; Long et al., 2003; Signor et al., 2016; Tanaka et al., 2009; Tanaka et al., 2011; Williams et al., 2008; Yassin et al., 2016). The needs of comparative research have in turn provided the impetus for molecular phylogenetic studies in order to establish a reliable basis for trait reconstruction. To date, many studies have examined the phylogeny of the melanogaster species group using overlapping datasets and taxon samples (Barmina and Kopp, 2007; Catullo and Oakeshott, 2014; Chen et al., 2014; Da Lage et al., 2007; Goto and Kimura, 2001; Kopp, 2006; Kopp and True, 2002; Lewis et al., 2005; Matsuda et al., 2009; Schawaroch, 2002).
In addition to the well-studied melanogaster, obscura, willistoni, and saltans species groups, Sophophora includes several smaller species groups as well as a number of unplaced species (O’Grady and DeSalle, 2018). In this report, we focus on the dentissima and fima lineages, both of which are endemic to Africa. The fima group occurs in both lowland and montaine habitats and displays a strong ecological specialization, breeding almost exclusively on native figs (Burla, 1954; Lachaise and Chassagnard, 2002; Tsacas and Lachaise, 1981). The dentissima group is restricted to highland forests; its ecological habits are less clear, but it appears to use a wider range of food sources (Lachaise and Chassagnard, 2001; Tsacas, 1980a, b). Aside from one study that included a single species, D. fima (Pelandakis et al., 1991), no members of the dentissima and fima lineages have been included in previous molecular phylogenies. Here, we show that both these lineages cluster within the broadly defined melanogaster species group. We discuss the implications of this result for Sophophora taxonomy, as well as for the evolution of male-specific morphological traits and ecological specialization.
2. Materials and Methods
2. 1. Drosophila specimens and species identification
A live strain of D. fima was kindly provided by Dr. J. R. David (CNRS, Gif-sur-Yvette, France). Most of the remaining fima group species were only available as fixed specimens collected by Dr. D. Lachaise and presented to us by J. R. David. Specimens with IDs starting with F and those starting with G came from different field collections, but the locations and dates of these collections are unknown, except that they originated from equatorial West Africa in the late 1970s or early 1980s. D. lamottei specimen B8 was collected by J. R. David at Mt. Oku in Cameroon. D. lamottei H1 and D. matilei specimen H3 were collected by sweeping by S. R. Prigent in the Oku Kilim forest, Cameroon, at 2200-2400m elevation, on December 18, 2012. D. microralis specimen H5 was collected by sweeping over fallen figs by S. R. Prigent in Oku Koh Kesoten, Cameroon, at 2100m elevation, on December 22, 2012.
Front legs from each male individual were dissected, mounted in Hoyers media between two coverslips, and photographed under brightfield illumination. The terminalia from the same individual were dissected, mounted, and used for species identification. The rest of each fly was used for DNA extraction and sequencing.
The fima and dentissima groups include 23 and 18 formally described species, respectively (Burla, 1954; Lachaise and Chassagnard, 2001, 2002; Tsacas, 1980a, b; Tsacas and Lachaise, 1981). Among the samples in hand, 9 species of the fima group and 3 species of the dentissima group were identified. Species are distinguishable by a combination of characters, including body coloration, the presence and morphology of male sex combs, and the shape of the phallic and periphallic organs. In the fima group, species can be partitioned into those without sex combs, those with simple sex combs, and those with double sex combs. Most species without sex combs were described by Burla (Burla, 1954), with one species added later by Lachaise and Chassagnard (Lachaise and Chassagnard, 2002). All the “sexcombless” species described by Burla (Burla, 1954) have similar periphallic organs, and their phallic organs have never been described. Their identification is therefore based mostly on pigmentation. However, careful examination of male genitalia allowed us to distinguish several species that were further identified by their color patterns as D. fima Burla, 1954 (dark notum and light pleurites), D. abure Burla, 1954 (entire thorax light yellow-brown), and D. kulango Burla, 1954 (entire thorax dark brown). The species with sex combs have received more attention, and the descriptions of their genitalia are more complete. Among the specimens with simple sex combs, we used male genital characters to identify D. dyula Burla, 1954, D. petitae Tsacas, 1981, D. microralis Tsacas, 1981 (also characterized by its blunt oral setae), and an undetermined species showing some similarities to D. sycophaga Tsacas, 1981. The two described species with double sex combs, D. akai and D. alladian, are easily separated and were both found among our samples.
The D. dentissima species group has been revised by Tsacas (Tsacas, 1980a, b), who established 5 species complexes and described the genitalia of all species known at the time. Later, three more species were added (Lachaise and Chassagnard, 2001; Tsacas, 1985). The specimens available to us belong to two species complexes according to their sex comb structure: the lamottei and matilei complexes. The observation of the genitalia allowed us to identify D. matilei Tsacas, 1974 (Tsacas, 1974), D. lamottei Tsacas, 1980, and D. anisoctena Tsacas, 1980. The last two species are closely related and show very similar genitalia, but can be distinguished by minor differences in the ventral part of the epandrium, the setation on the surstylus and the anal plate, and the shape of anterior and posterior parameres.
2. 2. DNA amplification and sequencing
The “F” and “G” collections have been stored at room temperature in ethanol of unknown concentration for several decades, and DNA preservation was found to be poor. DNA was extracted using an affinity resin based protocol (Hi Yield® Genomic DNA Mini Kit, Süd-Laborbedarf Gauting, Germany); several other DNA extraction methods tested on female specimens from the same collections produced similar results. For most specimens, two rounds of nested PCR were required to amplify nuclear loci in detectable amounts. PCR was carried out using DreamTaq polymerase (Thermo Fisher) and the following cycling conditions: 95° 5’ => (95° 30” => 55° 30” => 72° 80”)x35 => 72° 5’ =>12°; primer sequences are listed in Table S1. Amplified fragments were gel-purified and sequenced from both ends using amplification primers. Sequence chromatograms were examined and trimmed in SnapGene Viewer, then the two end reads were aligned and edited in Geneious (Kearse et al., 2012). Heterozygous nucleotide positions, if present, were represented by IUPAC ambiguity codes. All new sequences were deposited in Genbank under accession numbers listed in Table S2. Homologous sequences from species outside the fima and dentissima groups were obtained from Genbank or extracted from whole-genome sequences using Blast v2.2.23; the relevant accession numbers and sources of genome sequences are listed in Table S3.
2. 3. Sequence analysis
The sequences of each locus were aligned using the MUSCLE algorithm (Edgar, 2004) implemented in Geneious (Kearse et al., 2012). The alignments were trimmed at the ends, and poorly aligning intronic regions were removed. A gene tree was reconstructed from each locus separately using a heuristic maximum likelihood method implemented in PhyML 3.0 (Guindon et al., 2009; Guindon et al., 2005). All loci produced similar phylogenies with no strongly supported conflicting nodes (Figure S1). The alignments of all five loci were then concatenated for combined analysis. Combined Bayesian analysis was carried out in MrBayes v3.2.6 (Ronquist et al., 2012). Each locus was allowed to follow a different nucleotide substitution model with empirically estimated parameters. Two parallel runs of 1,500,000 generations, each starting from a different random tree, were carried out, and convergence was confirmed by comparing tree likelihoods and model parameters between the two runs. Trees were sampled every 1000 generations and summarized after a 20% relative burn-in. This analysis was repeated twice, producing identical consensus trees with posterior probabilities within 1% of each other at all nodes. Samples of probable trees were extracted from the .tprobs file, and a strict consensus of most probable trees with combined posterior probabilities of 95% or 99% was constructed from these sets of trees in Geneious. Consensus trees were then formatted using FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
3. Results
We have sequenced partial coding sequences of five nuclear, protein-coding loci: acon, eno, glyp, Pepck, and Pgm. In a maximum likelihood analysis of each locus separately, the fima species group was always monophyletic with strong support (Table 1, Figure S1). The separate analyses identified four clades within the fima group that were observed reproducibly and usually with strong support: (D. dyula + D. petitae), (D. microralis + D. aff. sycophaga), (D. fima + D. kulango + D. abure), and (D. alladian + D. akai). The relationships among these clades differed from locus to locus (Figure S1), but the (D. dyula + D. petitae) clade was almost always basal within the fima group (Table 1). Despite strong similarities between the sex combs of the (D. dyula + D. petitae) and (D. microralis + D. aff. sycophaga) clades, a sister-group relationship between these clades was not supported by any of the loci. The fima group as a whole always clustered within the melanogaster species group (sensu lato) with strong support, although its position within the melanogaster group varied among loci (Table 1, Figure S1).
Table 1.
Clade support from single-locus and combined maximum likelihood analyses.
| Clade | Species | Locus |
combined | ||||
|---|---|---|---|---|---|---|---|
| acon | eno | glyp | Pepck | Pgm | |||
| Clade 1 | D. dyula + D. petitae | 98 | 95 | 93 | 97 | -- | 100 |
| Clade 2 | D. microralis + D.. aff. sycophaga | 100 | 94 | 96 | 100 | 92 | 100 |
| Clade 3 | D. fima, D. kulango, D. abure | 98 | 97 | 87 | 98 | 99 | 100 |
| Clade 4 | D. alladian, D. akai | 100 | -- | 93 | 100 | 100 | 100 |
| Clade 1 most basal | 94 | -- | 95 | 94 | 100 | 100 | |
| Clades (1 + 2) sisters | -- | -- | -- | -- | -- | -- | |
| fima clade monophyletic | 100 | 100 | 99 | 100 | 100 | 100 | |
| (fima + dentissima) | -- | -- | -- | -- | -- | -- | |
| fima clade clustered within melanogaster species group | 100 | 98 | 100 | 100 | 98 | 100 | |
| dentissima clade within Oriental clade | 92 | 98 | 96 | 87 | 94 | 100 | |
| sister group to fima clade | ananassae subgroup | Oriental clade | ananassae subgroup | ananassae subgroup | ananassae subgroup | ananassae subgroup | |
| sister group to dentissima clade | D. elegans | D. elegans | D. ficusphila | D. elegans | (elegans + rhopaloa) subgroups | (elegans + rhopaloa) subgroups | |
Each cell shows the approximate likelihood ratio test statistic for the corresponding node (Guindon et al., 2009; Guindon et al., 2005). A dash indicates that the tree did not contain the indicated node. See Supplementary Figure S1 for single-locus trees, and Fig. S3 for the combined maximum likelihood tree.
The dentissima species group was also placed within the melanogaster species group (sensu lato), and more specifically within the “Oriental” clade (Kopp, 2006; Kopp and True, 2002), with strong support in the separate analysis of all five loci (Table 1). A sister-group relationship between the dentissima and fima groups was not observed for any of the loci (Table 1).
Since all loci agreed on the key aspects of the fima and dentissima group phylogeny, we concatenated the sequences of all five loci for a combined analysis. In the resulting Bayesian phylogeny, most relationships are supported with 100% posterior probabilities. The fima species group is monophyletic and contains all four clades that were identified in the separate analyses (Table 1), with the (D. dyula + D. petitae) clade most basal (Figure 1). This phylogeny provides strong evidence against a sister-group relationship between the (D. dyula + D. petitae) and (D. microralis + D. aff. sycophaga) clades. The fima species group is placed within the melanogaster species group (sensu lato), and more specifically as the sister lineage to the ananassae subgroup (sometimes considered a species group; see (Da Lage et al., 2007)), with 100% posterior probability. The dentissima group is placed within the Oriental clade, as the sister lineage to the (elegans + rhopaloa) subgroups, also with 100% posterior probability (Figure 1). A sister-group relationship between the dentissima and fima groups is not observed in any of the 28 Bayesian trees with posterior probabilities of 0.1% or higher.
Figure 1.
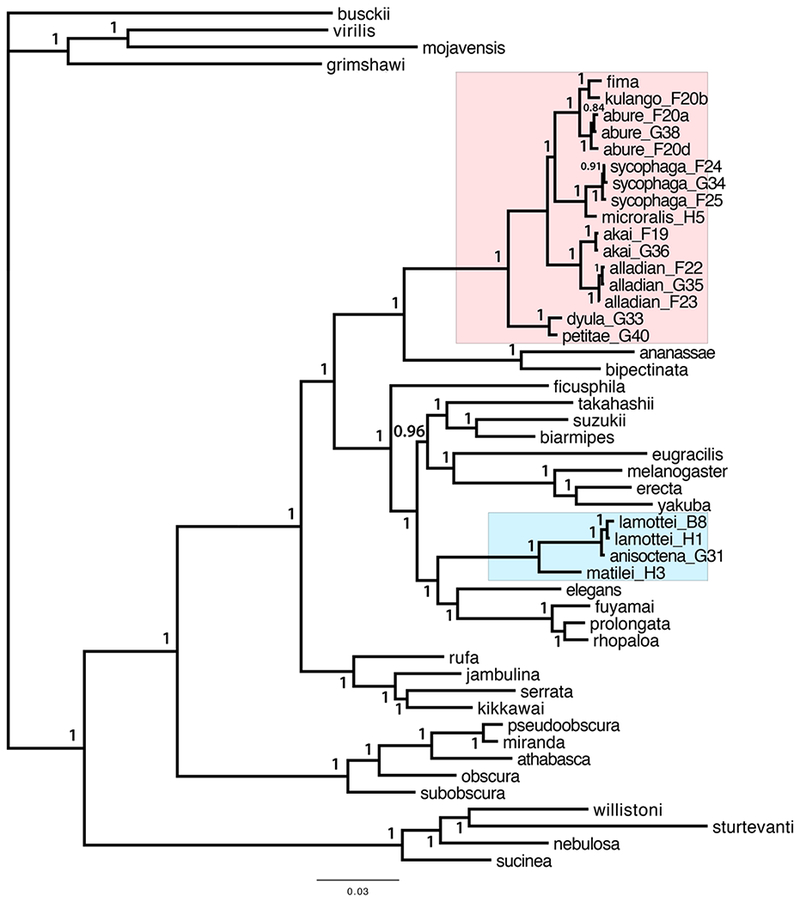
A. Bayesian phylogeny based on the combined 5-locus dataset. The fima clade is boxed in pink, and the dentissima clade in light blue. Letters and numbers following species names indicate specimen IDs (see Materials and Methods and Supplement Table S2). Taxonomic affiliations of other species are indicated in Supplement Table S3. Numbers at each node indicate the posterior probabilities of the respective taxon bipartitions.
We constructed a strict consensus of 16 trees with the cumulative posterior probability of 99%. In this tree, two important phylogenetic relationships are not fully resolved: namely, it is not certain whether the ananassae or the montium subgroup is the most basal within the melanogaster species group, and the relationships among several species subgroups within the Oriental clade are also unclear (Figure S2). These parts of the melanogaster group phylogeny were also the most difficult to resolve in previous analyses using different multilocus datasets (Barmina and Kopp, 2007; Chen et al., 2014; Da Lage et al., 2007; Kopp, 2006; Kopp and True, 2002; Schawaroch, 2002; Wong et al., 2007). However, the strict consensus tree has 100% support for placing both the fima and the dentissima groups within the melanogaster species group (sensu lato), for placing the fima group as sister to the ananassae subgroup, and for placing the dentissima group as sister to the (elegans + rhopaloa) subgroups (Figure S2). All these relationships are also observed in the strict consensus of 28 trees with the cumulative posterior probability of 99.9% (not shown). A combined analysis of all five loci using maximum likelihood supports the same key relationships, with approximate likelihood ratio test statistic = 100% for all relevant nodes (Figure S3). The maximum likelihood analysis also supports the same relationships within the fima species group as the Bayesian analysis (Figure 1, S3). Taking into account both the separate and the combined analyses, we can conclude that the fima and dentissima species groups are clades within the melanogaster species group (sensu lato), but are distantly related to each other.
4. Discussion
4.1. melanogaster species group and the taxonomy of Sophophora
Historically, drosophilists have utilized informal taxonomic ranks between the species and subgenus levels (lineage, radiation, group, subgroup, species complex) to better illustrate the continuum of species diversification (O’Grady and DeSalle, 2018). Similar to classical taxa, each new taxon was defined by morphological characters shared only by all included species. As stated by Bock and Wheeler (Bock and Wheeler, 1972), each species group was devised to illustrate a burst of speciation in a particular geographic region, and was supposed to contain closely related species, of similar morphology, with comparable chromosome banding patterns, and sometimes able to cross-hybridize under laboratory conditions. The species groups were originally erected to encompass small numbers of species. When Sturtevant (Sturtevant, 1942) established the melanogaster species group, it contained only 14 species.
The ananassae and montium lineages were originally classified as subgroups within the melanogaster species group (Bock, 1980; Bock and Wheeler, 1972; Hsu, 1949; Lemeunier et al., 1986; Patterson and Stone, 1952; Toda, 1991). As the number of described species continued to accumulate (reaching nearly 200 species at present), it became clear that this group was far larger than other Drosophila species groups, and encompassed exceptional levels of morphological diversity. In this sense, the melanogaster group has been moving steadily away from the original conception of a “species group”. The montium and ananassae subgroups, now including 94 and 27 species, respectively, were each confirmed to be monophyletic, and to form the two most basal lineages in the melanogaster species group, by multiple phylogenetic analyses (Barmina and Kopp, 2007; Chen et al., 2014; Da Lage et al., 2007; Kopp, 2006; Schawaroch, 2002). Thus, Da Lage et al (Da Lage et al., 2007) proposed to upgrade the ananassae and montium subgroups to species group status, splitting the original melanogaster group of Sturtevant (Sturtevant, 1942) into three species groups: ananassae, montium, and the reduced melanogaster group (sensu stricto) consisting of all the remaining subgroups (“the Oriental lineage” of Kopp and True (Kopp and True, 2002)). This classification has been followed by some authors (van der Linde et al., 2010; Yassin, 2013, 2018), but has not been widely adopted (Chen et al., 2014; Levine et al., 2016; McEvey and Schiffer, 2015; Tanaka et al., 2011; Watada et al., 2011).
The fima species group, proposed by Burla (Burla, 1954) and defined in more detail by Tsacas and Lachaise (Tsacas and Lachaise, 1981), was considered to be a member of Sophophora, but its position within the subgenus remained unclear. Burla (1954) noted a close relationship between the fima group and D. ananassae, but refrained from including D. ananassae in the fima group. The only phylogenetic study to address this question included a single representative species (D. fima) and was based on a single locus, 28S rRNA (Pelandakis et al., 1991). In that study, D. fima was placed as sister to the ananassae subgroup, but with low support. The dentissima lineage was originally proposed as a subgroup within the melanogaster species group by Bock and Wheeler (Bock and Wheeler, 1972), but was later upgraded to species group status by Tsacas (Tsacas, 1980a, b).
Our results clarify the phylogenetic relationships among different groups and subgroups, but leave several valid taxonomic options. A sister-group relationship between the fima species group and the ananassae group/subgroup is strongly supported in the combined analysis, as is the placement of the dentissima lineage within the Oriental clade (the reduced melanogaster species group sensu Da Lage et al (Da Lage et al., 2007)), and more specifically as sister to the (elegans + rhopaloa) subgroups. Thus, it is highly likely that both the fima and the dentissima lineages are contained within the original melanogaster species group sensu Sturtevant (Sturtevant, 1942) and Bock and Wheeler (Bock and Wheeler, 1972).
There are two obvious paths to preserving monophyletic species groups, each with its own advantages and disadvantages. First, if we adopt the taxonomy of Da Lage (Da Lage et al., 2007) and Yassin (Yassin, 2013, 2018), the ananassae, montium, and fima clades should be treated as separate species groups. For the dentissima clade, one of the options would be to include it as a subgroup within the narrowly defined melanogaster species group sensu Da Lage et al (Da Lage et al., 2007), along with the melanogaster, eugracilis, takahashii, ficusphila, elegans, rhopaloa, and favohirta subgroups (Bock, 1980). The advantage of this approach is that it keeps different species groups within Sophophora at roughly comparable sizes, and hews closer to the original conception of a species group as encompassing a modest amount of morphological diversity. The fima and dentissima lineages are well defined monophyletic taxa. They include, respectively, 23 and 18 formally described, closely related species, all endemic to Africa. In this sense, they fit the historical definition of the species-group taxonomic rank. Moreover, Burla (Burla, 1954) erected the fima group because its member species were morphologically too different from the diagnostic definition of the melanogaster group. Later, Tsacas and Lachaise (Tsacas and Lachaise, 1981) showed that the species of the fima group shared an unusual functioning of the male genitalia. Similarly, Tsacas (Tsacas, 1980b) discriminated the dentissima group from the melanogaster group based on peculiarities of the male genitalia.
The melanogaster group (sensu lato) already encompasses exceptional levels of morphological disparity, as discussed by Da Lage et al (Da Lage et al., 2007). Additional inclusion of the fima and dentissima lineages would further complicate the efforts to provide a concise diagnosis of the melanogaster species group. It is important to note that the subgenus Sophophora, as currently defined, is paraphyletic, as the willistoni and saltans species groups are more closely related to the Lordiphosa genus than to the melanogaster and obscura species groups (Gao et al., 2011). Maintaining a robust morphological definition of the melanogaster species group would probably make it easier to define broader monophyletic taxa within the overall (Sophophora + Lordiphosa) radiation. Based on our molecular phylogeny, it would be straightforward to treat the fima, ananassae, montium, and melanogaster (including dentissima) lineages as separate species groups, especially in light of the long and well-supported branches at the base of each of these groups. Establishing the dentissima lineage as a separate species group would pose greater challenges, since that would necessitate splitting the Oriental lineage sensu Kopp and True (Kopp and True, 2002) and the melanogaster group sensu stricto (Da Lage et al., 2007) into multiple species groups – an especially complicated proposition given that some of the relationships among subgroups within the Oriental lineage are still not entirely clear (Barmina and Kopp, 2007; Chen et al., 2014; Da Lage et al., 2007; Goto and Kimura, 2001; Kopp, 2006; Kopp and True, 2002; Schawaroch, 2002).
An alternative, and very different approach woud be to preserve the original taxonomy of Bock and Wheeler (Bock and Wheeler, 1972), with a broadly inclusive melanogaster species group sensu Sturtevant (Sturtevant, 1942). In this case, the fima group needs to be downgraded to subgroup status, and the ananassae, montium, fima, and dentissima clades are all to be considered species subgroups within the melanogaster group. This approach results in a rather inwieldy species group consisting of over 230 morphologically diverse species. This makes the “lumping” approach difficult to justify on purely taxonomic grounds. However, it has the important advantage of preserving continuity in the non-taxonomic literature. The melanogaster species group occupies a special place among Drosophila, in that the vast majority of the literature on this clade is composed of comparative studies rather than phylogenetic or taxonomic ones. Dozens of publications that use D. melanogaster and its relatives as models for molecular evolution, comparative genomics, and evolutionary developmental biology refer to species from the ananassae and montium clades as members of the melanogaster species group (Atallah et al., 2012; Barmina and Kopp, 2007; Chen et al., 2010; Chen et al., 2014; Jeong et al., 2006; Kursel and Malik, 2017; Levine et al., 2012; Levine et al., 2016; Long et al., 2003; Signor et al., 2016; Tanaka et al., 2009; Tanaka et al., 2011; Williams et al., 2008; Yassin et al., 2016). Removing these clades from the melanogaster group can lead to confusion if different authors, or same authors writing at different times, include narrower vs wider sets of species under the name “melanogaster species group”. Precluding such potential confusion is a strong reason for taxonomic conservatism despite its obvious disadvantages.
We also note that several lineages traditionally included in the melanogaster species group, for example the denticulata and longissima subgroups (Bock, 1980; Bock and Wheeler, 1972; Toda, 1991), or D. pinnitarsus and its close relatives (Bock, 1976), are yet to be included in any molecular phylogenies. D. setifemur (syn. D. dispar; (McEvey, 2009)) was originally described as the type species of the dispar species group within Sophophora (Mather, 1955), and considered “not closely related to any other sophophoran species” (Bock and Parsons, 1978). However, a phylogenetic study has placed D. setifemur within the melanogaster species group, as sister to the ananassae subgroup (Barmina and Kopp, 2007); its position relative to the fima clade is yet to be determined. The position of D. ironensis, an unusual species tentatively assigned to the ananassae subgroup (Bock and Parsons, 1978), is also unclear; preliminary evidence suggests that it falls outside of the traditionally defined ananassae subgroup (Kopp, unpublished). Depending on the eventual placement of these and several other taxa, the taxonomy of the melanogaster species group (sensu lato) may need to undergo further revisions, which may be easier to accommodate and cause fewer back-and-forth changes if the melanogaster group continues to be defined broadly sensu Sturtevant (Sturtevant, 1942) and Bock and Wheeler (Bock and Wheeler, 1972). On balance, a conservative approach may be preferable until the full phylogeny of this lineage is determined.
4. 2. Species complexes in the fima clade
Tsacas and Lachaise (Tsacas and Lachaise, 1981) have subdivided the fima species group into three sets of species (“ensembles”): those with “anterior tarsi of males normal, without sex combs” (including D. fima, D. abure, and D. kulango), those with “anterior tarsi of males normal but with two sex combs on the first two segments” (D. alladian and D. akai), and those with “anterior tarsi of males with a modified second segment carrying a dorsal extension with a small transverse comb”. This third set of species, which is the largest in the fima clade, was later named the dyula species complex (Burla, 1954; Chassagnard et al., 1997; Lachaise and Chassagnard, 2002). However, this set of species can be further subdivided in two according to the aedeagus morphology (Tsacas and Lachaise, 1981). The species with glabrous aedeagus that rotates for erection (including D. dyula and D. petitae) are supposed to be the most derived with respect to genital function. The other species (including D. microralis and D. aff. sycophaga) have aedeagus that is hairy at the tip and does not rotate for erection; this is similar to D. fima, which belongs to the “sexcombless” set of species. In our phylogeny, the “dyula species complex” is clearly polyphyletic, as there is strong evidence that the (D. dyula + D. petitae) and (D. microralis + D. aff. sycophaga) clades are distantly related. This separation is consistent with the differences in male genital traits, namely with the presence or absence of hairs on the tip of the aedeagus and setation of the epandrium. In contrast, the species without sex combs, and those with double sex combs, form well-supported monophyletic clades in our analysis, although our taxon sampling is obviously limited.
Thus, our results suggest the subdivision of the fima lineage into four species complexes: the narrowly defined dyula complex, which diverges first within the fima clade and is restricted to the species with glabrous aedeagus; the akai complex, which diverges next and includes the species with double combs on each of the first two tarsomeres; the fima complex, including species without sex combs; and the sycophaga complex, consisting of the species that were previously included in the dyula complex but have hairy tip of the phallus. In light of the close relationship between the fima and sycophaga species complexes, the position of D. inopinata, a species that lacks sex combs but otherwise seems to belong to the sycophaga complex, deserves examination.
4. 3. Sex comb origin, diversification, and secondary loss
Male sex combs, which develop from modified mechanosensory bristles on the tarsal segments of front legs, are present in Old World Sophophora (the melanogaster, obscura, and fima clades) (reviewed in Kopp (Kopp, 2011)) and in some species of the genus Lordiphosa (Fartyal et al., 2017; Gao et al., 2011; Hu and Toda, 2000; Katoh et al., 2018; Lastovka and Maca, 1978; Lee, 1959). The subgenus Sophophora as established by Sturtevant (Sturtevant, 1939; Sturtevant, 1942) is in fact paraphyletic, since the New World Sophophora (the willistoni and saltans species groups, which lack sex combs) are more closely related to Lordiphosa than to Old World Sophophora (Gao et al., 2011). It is not clear whether the presence of sex combs in some Lordiphosa reflects a single origin of sex combs at the base of (Sophophora + Lordiphosa) followed by secondary loss in New World Sophophora and in most Lordiphosa species, or independent origin of sex combs in Old World Sophophora and in some species groups within Lordiphosa. The cellular mechanisms of sex comb development differ between the melanogaster and obscura species groups on the one hand, and the miki species group of Lordiphosa (Atallah et al., 2012). However, the cell biology of sex comb development varies just as drastically within the melanogaster species group (Tanaka et al., 2009), so no firm conclusions can be drawn on this basis. Both in the Old World Sophophora and in Lordiphosa, sex comb morphology is highy diverse, showing many examples of rapid divergence, convergent evolution, and secondary loss (Katoh et al., 2018; Kopp, 2011; Lemeunier et al., 1986; Tanaka et al., 2011).
Our results show that the fima and dentissima clades fall within the Old World Sophophora radiation, so their sex combs clearly share a single origin with those of the melanogaster and obscura species groups. Unfortunately, no species except D. fima are currently available in culture, preventing us from investigating sex comb development in the fima and dentissima clades. Within the fima clade, our phylogeny (Fig. 1) strongly suggests that species of the fima species complex (including D. fima, D. abure, D. kulango, and probably other similar species such as D. abron and D. iroko) have most likely lost sex combs secondarily. However, vestigial transverse sex combs are in fact present in D. abure (Figure 2), which may reflect an intermediate stage in the loss of sex combs. Similar transverse sex combs are found in the ananassae, takahashii, and elegans subgroups, as well as in the Lordiphosa denticeps species group (Katoh et al., 2018; Kopp, 2011). The robust, obliquely rotated sex combs of the akai species complex (D. alladian and D. akai) (Figure 2) strongly resemble the sex combs found in such melanogaster group species as D. biarmipes, D. suzukii, and the rhopaloa species subgroup in the Oriental clade, and D. bipectinata and D. parabipectinata in the ananassae subgroup. We may speculate that these sex combs develop through similar cellular mechanisms, i.e. by active epithelial rotation (Tanaka et al., 2009). Regardless of the mechanism, the diversity of sex comb morphologies in the fima clade reinforces the general pattern of rapid divergence, convergent evolution, and recurrent secondary loss (Kopp, 2011).
Figure 2.

Sex comb diversity in the Drosophila fima clade. The two most proximal tarsal segments (ta1 and ta2) of the male front leg are shown for each species. A. D. fima. B. D. kulango. C. D. abure. Note the vestigial transverse sex comb at the distal tip of ta2 (arrowhead). D. D. aff. sycophaga. E. D. microralis. F. D. alladian. G. D. akai. H. D. dyula. I. D. petitae. In D. aff. sycophaga, D. microralis, D. dyula, and D. petitae, sex combs are located on distal outgrowths of the ta1 and ta2 segments (arrowheads in D).
Transverse sex combs associated with robust distal outgrowths of the first and especially second tarsal segments are found in many species of the fima lineage (Lachaise and Chassagnard, 2002; Tsacas and Lachaise, 1981) (Figure 2). Our analysis suggests that these species belong to two distantly related clades: the dyula species complex, represented by D. dyula and D. petitae, and the sycophaga species complex, represented by D. microralis and D. aff. sycophaga (Figure 1). It is possible that this type of sex comb was present in the last common ancestor of the fima lineage. Transverse sex combs located distally on the first and second tarsal segments, although without any tarsal outgrowths, are the primitive condition for the ananassae subgroup (Matsuda et al., 2009), suggesting that transverse sex combs were present in the last common ancestor of the (fima + ananassae) lineage, while the tarsal outgrowths are a fima lineage synapomorphy. Among more distant relatives, however, tarsal outgrowths at the base of the sex comb are seen in the ficusphila subgroup and in D. denticulata. Even larger tarsal outgrowths are found in males of some Hawaiian Drosophila species (Stark and O’Grady, 2009), which are very distantly related to Sophophora. Thus, male-specific tarsal outgrowths associated with sex combs and other types of modified bristles represent a yet another example of morphological convergence.
As noted by Tsacas (Tsacas, 1980a, b), different types of sex combs are observed in the dentissima group. The sex comb of D. matilei is very similar to that of D. ficusphila, despite the distant relationship between these species. This similarity extends to the presence of a second sparse row of thinner, pointed teeth located ventrally from the main, densely packed comb composed of thick blunt teeth (Figure 3). The sex combs of D. lamottei and D. anisoctena are more similar to those of the montium and rhopaloa species subgroups, and lack the secondary row of teeth. Despite their morphological similarities in adults, the sex combs of the montium and rhopaloa subgroups develop through very different cellular mechanisms (Tanaka et al., 2009). Based on the adult morphology, we cannot discern the mode of sex comb development in D. lamottei and D. anisoctena. However, given their closer relationship to the rhopaloa subgroup, we may speculate that their sex combs develop by active epithelial rotation, as they do in the rhopaloa subgroup (Tanaka et al., 2009). One very unusual aspect of leg morphology in D. lamottei and D. anisoctena (and in the rest of the lamottei species complex sensu Tsacas) is the extreme shortening of the first tarsal segment in males (Figure 3). Tsacas (Tsacas, 1980a, b) has suggested that this is a derived trait within the dentissima group; the close relationship between this subgroup and the (elegans + rhopaloa) lineage, which has tarsal segments of normal length, supports this view, but more extensive taxon sampling is required to confirm this.
Figure 3.
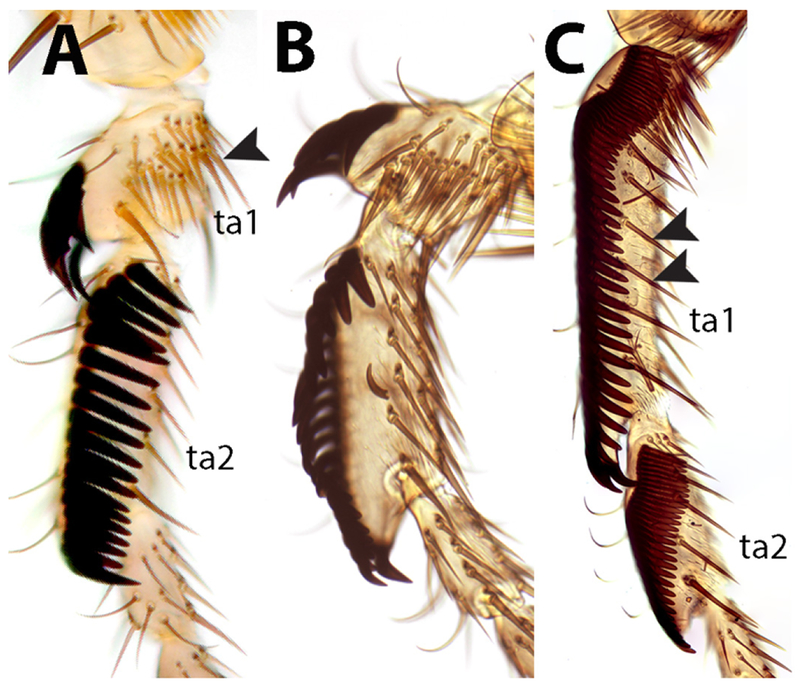
Sex comb diversity in the Drosophila dentissima clade. The two most proximal tarsal segments (ta1 and ta2) of the male front leg are shown for each species. A. D. lamottei. B. D. anisoctena. Note the extreme shortening of ta1 (arrowhead) in these two species. C. D. matilei. In addition to the main sex comb consisting of ~55 teeth on ta1 and ~30 teeth on ta2, a sparse longitudinal row of straight bristles is present more ventrally (double arrowheads).
4. 4. Multiple origins of fig-breeding
All 19 fima clade species whose ecological habits are known are associated with figs (Ficus sp.), breeding in the fig syconia (fruit) at various stages of decay (Lachaise and Chassagnard, 2002). Different species vary in the degree of their host plant specificity, but most exploit multiple fig species. Overall, this clade has been recorded from 16 species of Ficus, and some host species can harbor up to 12 fima-clade species (Lachaise and Chassagnard, 2002). Their fig specialization appears to be nearly complete, and is likely a synapomorphy of the fima clade: only D. fima and D. abron have been raised successfully on artificial media (Lachaise and Chassagnard, 2002). In the melanogaster species group, D. ficusphila also “has a special fondness for the fruits belonging to the genus Ficus such as F. carica and F. erecta” (Kikkawa and Peng, 1938), but can be easily raised on standard media. Outside Sophophora, the Afrotropical species of Lissocephala, another drosophilid genus, also appear to be strict fig specialists (Harry et al., 1996; Lachaise, 1977; Lachaise et al., 1982). Repeated evolution of fig specialization is perhaps not surprising, given the great abundance of this resource in many habitats – but interestingly, Oriental and Australasian species of Lissocephala do not specialize on figs despite the great abundance of Ficus species in those regions. Overall, the ecological and genetic factors favoring fig specialization over fruit generalism in Drosophilidae remain unknown.
Supplementary Material
Supplemental Figure 1. Maximum likelihood phylogenies based on individual loci. A. acon. B. eno. C. glyp. D. Pepck. E. Pgm. Numbers at each node indicate the approximate likelihood ratio test support for the respective clade. Support for clades of interest is summarized in Table 1. Locus and primer information can be found in Supplement Table S1, and Genbank accession numbers in Tables S2 and S3.
Supplemental Figure 2. B. Strict consensus of 16 trees with the cumulative posterior probability of 99%, labeled as in Figure 1. Numbers at each node indicate the posterior probabilities of the respective taxon bipartitions.
Supplemental Figure 3. Maximum likelihood phylogeny based on the combined 5-locus dataset, labeled as in Figure 1. Numbers at each node indicate the approximate likelihood ratio test support for the respective clade.
Highlights.
melanogaster species group in the largest and most diverse in Drosophila
fima and dentissima groups cluster within the melanogaster group sensu lato
we discuss the implications of this result for Drosophila taxonomy
male sex combs show many examples of convergent evolution
fig specialization has evolved multiple times in Drosophila
Acknowledgements
We are grateful to the late Daniel Lachaise for collecting most of the specimens used in this study, and to Jean R. David for sharing these specimens as well as the live strain of D. fima. We also thank Doris Bachtrog, Ryan Bracewell, Michael Bronski, Will Conner, Brandon Cooper, Danny Miller, Benjamin Prud’homme, and Junichi Yamaguchi for sharing unpublished genome and transcriptome assemblies of D. athabasca, D. fuyamai, D. jambulina, D. nebulosa, D. rufa, D. sturtevanti, and D. succinea, and Amir Yassin for extensive comments on the manuscript. AK and OB wish to thank Nicolas Gompel for allowing much of this work to be conducted in his lab while on a sabbatical visit, and to members of the Gompel lab for treating them as their own labmates. This work was supported by NIH grant 5R35GM122592 to AK and by funds from Ludwig-Maximilians-Universität München to Nicolas Gompel.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atallah J, Watabe H, and Kopp A (2012). Many ways to make a novel structure: a new mode of sex comb development in Drosophilidae. Evol Dev 14, 476–483. [DOI] [PubMed] [Google Scholar]
- Barmina O, and Kopp A (2007). Sex-specific expression of a HOX gene associated with rapid morphological evolution. Dev Biol 311, 277–286. [DOI] [PubMed] [Google Scholar]
- Bock IR (1976). Drosophilidae of Australia I. Drosophila (Insecta: Diptera). Australian Journal of Zoology 40, 1–105. [Google Scholar]
- Bock IR (1980). Current status of the Drosophila melanogaster species-group. (Diptera). Syst Ent 5, 341–356. [Google Scholar]
- Bock IR, and Parsons PA (1978). Australian Endemic Drosophila IV. Queensland Rain Forest Species Collected at Fruit Baits, with Descriptions of Two Species. Aust J Zool 26, 91–103. [Google Scholar]
- Bock IR, and Wheeler MR (1972). The Drosophila melanogaster species group. Univ Texas Publs 7, 1–102. [Google Scholar]
- Burla H (1954). Zur Kenntnis der Drosophiliden der Elfenbeinkuste (Französisch West-Afrika). Revue Suisse de Zoologie 61 (Supplement), 1–218. [Google Scholar]
- Catullo RA, and Oakeshott JG (2014). Problems with data quality in the reconstruction of evolutionary relationships in the Drosophila melanogaster species group: Comments on Yang et al. (2012). Mol Phylogenet Evol 78, 275–276. [DOI] [PubMed] [Google Scholar]
- Chassagnard M-T, Tsacas L, and Lachaise D (1997). Drosophilidae (Diptera) of Malawi. Ann Natal Mus 38, 61–131. [Google Scholar]
- Chen S, Zhang YE, and Long M (2010). New genes in Drosophila quickly become essential. Science 330, 1682–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZX, Sturgill D, Qu J, Jiang H, Park S, Boley N, Suzuki AM, Fletcher AR, Plachetzki DC, FitzGerald PC, et al. (2014). Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Res 24, 1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Lage J-L, Kergoat GJ, Maczkowiak F, Silvain J-F, Cariou M-L, and Lachaise D (2007). A phylogeny of Drosophilidae using the Amyrel gene: questioning the Drosophila melanogaster species group boundaries. J Zool Syst Evol Res 45, 47–63. [Google Scholar]
- Edgar RC (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fartyal RS, Sati PC, Pradhan S, Kandpal MC, Toda MJ, Chatterjee RN, Singh BK, and Bhardwai A (2017). A review of the genus Lordiphosa Basden in India, with descriptions of four new species from the Himalayan region (Diptera, Drosophilidae). Zookeys, 49–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao JJ, Hu YG, Toda MJ, Katoh T, and Tamura K (2011). Phylogenetic relationships between Sophophora and Lordiphosa, with proposition of a hypothesis on the vicariant divergences of tropical lineages between the Old and New Worlds in the family Drosophilidae. Mol Phylogenet Evol 60, 98–107. [DOI] [PubMed] [Google Scholar]
- Gao JJ, Watabe HA, Toda MJ, Zhang YP, and Aotsuka T (2003). The Drosophila obscura species-group (Diptera, Drosophilidae) from Yunnan Province, Southern China. Zoolog Sci 20, 773–782. [DOI] [PubMed] [Google Scholar]
- Gleason JM, Griffith EC, and Powell JR (1998). A Molecular Phylogeny of the Drosophila Willistoni Group: Conflicts between Species Concepts? Evolution 52, 1093–1103. [DOI] [PubMed] [Google Scholar]
- Goto SG, and Kimura MT (2001). Phylogenetic utility of mitochondrial COI and nuclear Gpdh genes in Drosophila. Mol Phylogenet Evol 18, 404–422. [DOI] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, and Gascuel O (2009). Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol 537, 113–137. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, and Gascuel O (2005). PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33, W557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry M, Solignac M, and Lachaise D (1996). Adaptive radiation in the Afrotropical region of the Paleotropical genus Lissocephala (Drosophilidae) on the pantropical genus Ficus (Moraceae). Journal of Biogeography 23, 543–552. [Google Scholar]
- Hsu TC (1949). The external genital apparatus of male Drosophilidae in relation to systematics. Univ Texas Publs 4920, 80–142. [Google Scholar]
- Hu Y-G, and Toda MJ (2000). Polyphyly of Lordiphosa and its relationships in Drosophilinae (Diptera: Drosophilidae). Systematic Entomology 25, 1–17. [Google Scholar]
- Jeong S, Rokas A, and Carroll SB (2006). Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 125, 1387–1399. [DOI] [PubMed] [Google Scholar]
- Katoh T, Tamura K, and Aotsuka T (2000). Phylogenetic position of the subgenus Lordiphosa of the genus Drosophila (Diptera: Drosophilidae) inferred from alcohol dehydrogenase (Adh) gene sequences. J Mol Evol 51, 122–130. [DOI] [PubMed] [Google Scholar]
- Katoh TK, Zhang G, Toda MJ, Zhang WX, and Gao JJ (2018). The Lordiphosa denticeps species group (Diptera: Drosophilidae) in China, with redescriptions of four known species and descriptions of nine new species. Zootaxa 4471, 37–75. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa H, and Peng FT (1938). Drosophila species of Japan and adjacent localities. Japanese Journal of Zoology 7, 507–552. [Google Scholar]
- Kopp A (2006). Basal relationships in the Drosophila melanogaster species group. Mol Phylogenet Evol 39, 787–798. [DOI] [PubMed] [Google Scholar]
- Kopp A (2011). Drosophila Sex Combs as a Model of Evolutionary Innovations. Evol Dev 13, 504–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A, and True JR (2002). Phylogeny of the Oriental Drosophila melanogaster Species Group: A Multilocus Reconstruction. Syst Biol 51, 786–805. [DOI] [PubMed] [Google Scholar]
- Kursel LE, and Malik HS (2017). Recurrent Gene Duplication Leads to Diverse Repertoires of Centromeric Histones in Drosophila Species. Mol Biol Evol 34, 1445–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D (1977). Niche separation of African Lissocephala within the Ficus Drosophilid community. Oecologia 31, 201–214. [DOI] [PubMed] [Google Scholar]
- Lachaise D, and Chassagnard MT (2001). Seven new montane species of Drosophila in the Eastern Arc mountains and Mt Kilimanjaro in Tanzania attesting to past connections between eastern and western African mountains (Diptera: Drosophilidae). Eur J Entomol 98, 351–366. [Google Scholar]
- Lachaise D, and Chassagnard MT (2002). Divergence of sex comb phenotypes in the Drosophila fima species group and radiation on Afrotropical Ficus, including five new species from East Africa and Madagascar (Diptera: Drosophilidae). Annales de la Societe entomologique de France 38, 79–99. [Google Scholar]
- Lachaise D, Tsacas L, and Couturier G (1982). The Drosophilidae associated with tropical African figs. Evolution 36, 141–151. [DOI] [PubMed] [Google Scholar]
- Lakovaara S, and Saura A (1982). Evolution and speciation in the Drosophila obscura group In The Genetics and Biology of Drosophila, Ashburner M, Carson HL, and Thompson JN, eds. (London: Academic Press; ), pp. 1–59. [Google Scholar]
- Lastovka P, and Maca J (1978). European species of the Drosophila subgenus Lordiphosa (Diptera, Drosophilidae). Acta ent bohemoslovaca 75, 404–420. [Google Scholar]
- Lee TJ (1959). On a new species, “Drosophila clarofinis” sp. nov. Korean Journal of Zoology 2, 7–9. [Google Scholar]
- Lemeunier F, David JR, Tsacas L, and Ashburner M (1986). The melanogaster species group. In The Genetics and Biology of Drosophila, Ashburner M, Carson HL, and Thompson JN, eds. (New York, NY: Academic Press; ), pp. 147–256. [Google Scholar]
- Levine MT, McCoy C, Vermaak D, Lee YC, Hiatt MA, Matsen FA, and Malik HS (2012). Phylogenomic analysis reveals dynamic evolutionary history of the Drosophila heterochromatin protein 1 (HP1) gene family. PLoS Genet 8, e1002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MT, Vander Wende HM, Hsieh E, Baker EP, and Malik HS (2016). Recurrent Gene Duplication Diversifies Genome Defense Repertoire in Drosophila. Mol Biol Evol 33, 1641–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RL, Beckenbach AT, and Mooers AO (2005). The phylogeny of the subgroups within the melanogaster species group: Likelihood tests on COI and COII sequences and a Bayesian estimate of phylogeny. Mol Phylogenet Evol 37, 15–24. [DOI] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, and Wang W (2003). The origin of new genes: glimpses from the young and old. Nat Rev Genet 4, 865–875. [DOI] [PubMed] [Google Scholar]
- Mather WB (1955). The genus Drosophila (Diptera) in eastern Queensland. I. Taxonomy. Australian Journal of Zoology 3, 545–582. [Google Scholar]
- Matsuda M, Ng C-S, Doi M, Kopp A, and Tobari YN (2009). Evolution in the Drosophila ananassae species subgroup. Fly 3, 1–13. [DOI] [PubMed] [Google Scholar]
- McEvey SF (2009). Taxonomic Review of the Australian Drosophila setifemur Species Group, a New Name for the D. dispar Species Group (Diptera: Drosophilidae). Records of the Australian Museum 61, 31–38. [Google Scholar]
- McEvey SF, and Schiffer M (2015). New Species in the Drosophila ananassae Subgroup from Northern Australia, New Guinea and the South Pacific (Diptera: Drosophilidae), with Historical Overview. Records of the Australian Museum 67, 129–161. [Google Scholar]
- O’Grady PM (1999). Reevaluation of phylogeny in the Drosophila obscura species group based on combined analysis of nucleotide sequences. Mol Phylogenet Evol 12, 124–139. [DOI] [PubMed] [Google Scholar]
- O’Grady PM (2010). Whither Drosophila? Genetics 185, 703–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Grady PM, Clark JB, and Kidwell MG (1998). Phylogeny of the Drosophila saltans species group based on combined analysis of nuclear and mitochondrial DNA sequences. Mol Biol Evol 15, 656–664. [DOI] [PubMed] [Google Scholar]
- O’Grady PM, and DeSalle R (2018). Phylogeny of the Genus Drosophila. Genetics 209, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Grady PM, and Kidwell MG (2002). Phylogeny of the subgenus sophophora (Diptera: drosophilidae) based on combined analysis of nuclear and mitochondrial sequences. Mol Phylogenet Evol 22, 442–453. [DOI] [PubMed] [Google Scholar]
- O’Grady PM, and Markow TA (2009). Phylogenetic taxonomy in Drosophila. Fly 3, 10–14. [DOI] [PubMed] [Google Scholar]
- Patterson JT, and Stone WS (1952). Evolution in the genus Drosophila. (New York:Macmillan; ). [Google Scholar]
- Pelandakis M, Higgins DG, and Solignac M (1991). Molecular phylogeny of the subgenus Sophophora of Drosophila derived from large subunit of ribosomal RNA sequences. Genetica 84, 87–94. [DOI] [PubMed] [Google Scholar]
- Remsen J, and O’Grady P (2002). Phylogeny of Drosophilinae (Diptera: Drosophilidae), with comments on combined analysis and character support. Mol Phylogenet Evol 24, 249–264. [DOI] [PubMed] [Google Scholar]
- Robe LJ, Cordeiro J, Loreto EL, and Valente VL (2010). Taxonomic boundaries, phylogenetic relationships and biogeography of the Drosophila willistoni subgroup (Diptera: Drosophilidae). Genetica 138, 601–617. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Tarrio R, and Ayala FJ (1999). Molecular evolution an d phylogeny of the Drosophila saltans species group inferred from the Xdh gene. Mol Phylogenet Evol 13, 110–121. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, and Huelsenbeck JP (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61, 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schawaroch V (2002). Phylogeny of a paradigm lineage: the Drosophila melanogaster species group (Diptera : Drosophilidae). Biol J Linn Soc 76, 21–37. [Google Scholar]
- Signor SA, Liu Y, Rebeiz M, and Kopp A (2016). Genetic Convergence in the Evolution of Male-Specific Color Patterns in Drosophila. Curr Biol 26, 2423–2433. [DOI] [PubMed] [Google Scholar]
- Stark JB, and O’Grady PM (2009). Morphological variation in the forelegs of the Hawaiian Drosophilidae. I. The AMC clade. J Morphol. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH (1939). On the subdivision of the genus Drosophila. Proc Natl Acad Sci U S A 25, 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH (1942). The classification of the genus Drosophila, with descriptions of nine new species. Univ Texas Publs 4231, 6–51. [Google Scholar]
- Tanaka K, Barmina O, and Kopp A (2009). Distinct developmental mechanisms underlie the evolutionary diversification of Drosophila sex combs. Proceedings of the National Academy of Sciences 106, 4764–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Barmina O, Sanders LE, Arbeitman MN, and Kopp A (2011). Evolution of Sex-Specific Traits through Changes in HOX-Dependent doublesex Expression. PLoS Biol 9, e1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda MJ (1991). Drosophilidae (Diptera) in Myanmar (Burma) VII. The Drosophila melanogaster species-group, excepting the D. montium subgroup. Oriental Insects 25, 69–94. [Google Scholar]
- Tsacas L (1974). Drosophila matilei, nouvelle espèce de l’ouest Cameroun, du groupe melanogaster, et redescription de D. microlabis Séguy (Dipt. Drosophilidae). Bull Soc ent Fr 79, 145–151. [Google Scholar]
- Tsacas L (1980a). Les espèces montagnardes afrotropicales de Drosophilidae (Diptera) I - Le groupe Drosophila dentissima. Annales de la Société entomologique de France 16, 517–540. [Google Scholar]
- Tsacas L (1980b). Les groupes d’espèces du sous-genre Sophophora Sturtevant (Diptera, Drosophilidae, Drosophila) et le rôle du fonctionnement des génitalias males dans la définition des taxons supraspécifiques. Extrait du Bulletin de la Société Zoologique de France 105, 529–543. [Google Scholar]
- Tsacas L (1985). Drosophila desavrilia, n. sp. des montagnes malgaches {Diptera, Drosophilidae}. Revue fr Ent 7, 189–191. [Google Scholar]
- Tsacas L, and Lachaise D (1981). Les espèces au second article tarsal modifié du groupe afrotropical Drosophila fima (Diptera, Drosophilidae). Annales de la Société entomologique de France 17, 395–415. [Google Scholar]
- van der Linde K, Houle D, Spicer GS, and Steppan SJ (2010). A supermatrix-based molecular phylogeny of the family Drosophilidae. Genet Res (Camb) 92, 25–38. [DOI] [PubMed] [Google Scholar]
- Watada M, Matsumoto M, Kondo M, and Kimura M (2011). Taxonomic study of the Drosophila auraria species complex (Diptera: Drosophilidae) with description of a new species. Entomological Science 14, 392–398. [Google Scholar]
- Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, and Carroll SB (2008). The regulation and evolution of a genetic switch controlling sexually dimorph ic traits in Drosophila. Cell 134, 610–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Jensen JD, Pool JE, and Aquadro CF (2007). Phylogenetic incongruence in the Drosophila melanogaster species group. Mol Phylogenet Evol 43, 1138–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin A (2013). Phylogenetic classification of the Drosophilidae Rondani (Diptera): the role of morphology in the postgenomic era. Systematic Entomology 38, 349–364. [Google Scholar]
- Yassin A (2018). Phylogenetic biogeography and classification of the Drosophila montium species group (Diptera: Drosophilidae),. Annales de la Societe entomologique de France 54, 167–175. [Google Scholar]
- Yassin A, Delaney EK, Reddiex AJ, Seher TD, Bastide H, Appleton NC, Lack JB, David JR, Chenoweth SF, Pool JE, et al. (2016). The pdm3 Locus Is a Hotspot for Recurrent Evolution of Female-Limited Color Dimorphism in Drosophila. Curr Biol 26, 2412–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Maximum likelihood phylogenies based on individual loci. A. acon. B. eno. C. glyp. D. Pepck. E. Pgm. Numbers at each node indicate the approximate likelihood ratio test support for the respective clade. Support for clades of interest is summarized in Table 1. Locus and primer information can be found in Supplement Table S1, and Genbank accession numbers in Tables S2 and S3.
Supplemental Figure 2. B. Strict consensus of 16 trees with the cumulative posterior probability of 99%, labeled as in Figure 1. Numbers at each node indicate the posterior probabilities of the respective taxon bipartitions.
Supplemental Figure 3. Maximum likelihood phylogeny based on the combined 5-locus dataset, labeled as in Figure 1. Numbers at each node indicate the approximate likelihood ratio test support for the respective clade.


