Abstract
Mood disorders such as anxiety and depression are heterogeneous disorders with many sufferers unresponsive to current pharmacological treatments. Individual differences in temperament represent one factor that may underlie symptom heterogeneity, so understanding its biological underpinnings can help pave the way to personalized therapies and improved patient outcomes. The present study uses a rodent model of temperamental differences to examine whether individual differences in emotional behavior phenotypes correspond to altered limbic brain cellular metabolism, an indicator of neuronal activity. The model uses two selectively bred rat strains -- high novelty responder rats (HRs) that show highly exploratory behavior in a novel environment, active coping style and resilience to chronic mild stress compared to low novelty responder rats (LRs), which are inhibited in novel environments, display passive coping style, and are susceptible to chronic stress. Utilizing transcriptome data from a prior study in adult HR/LR rats, we first show that a preponderance of genes differing in the HR vs. LR hippocampus and amygdala are involved in cellular metabolism. This led us to then ask if oxygen consumption was altered in isolated mitochondria of the hippocampus and amygdala of HR/LR rats; here we found increased oxygen consumption reserve capacity in LR amygdala. Our last experiment examined activity of cytochrome c oxidase (COX), an enzyme responsible for ATP production and correlate of metabolic activity, in several brain regions of HR/LR rats. We found that LRs displayed higher COX activity in the dentate gyrus, prefrontal cortex, and dorsal raphe compared to HRs, with no significant HR/LR difference in nuclei of the amygdala. Correlational analyses of COX activity across brain regions suggested divergent connectivity between the prefrontal cortex, amygdala, hippocampus, and dorsal raphe of HR vs. LR rats. Together these studies point to altered cellular metabolism in the limbic brain of LR/HR animals, which may reflect altered neural circuitry that drives their divergent behavioral profiles.
Keywords: anxiety, transcriptome, metabolism, hippocampus, amygdala, cytochrome c oxidase
Section 1. Introduction
Vulnerability to mood disorders stems from a combination of environmental factors and innate biological factors, including those that drive individual differences in temperament and personality. Temperament is observable across species and can be useful method to study emotional behavior in model organisms. One way that temperamental differences can be measured is through one’s response to a novel environment (Reale et al., 2007). In humans, individuals with a low behavioral response to novelty (i.e., those who are inhibited in a new environment) are predisposed to mood disorders such as anxiety and depression (Celikel et al., 2009). On the other hand, individuals that show high novelty seeking (i.e., those who readily explore new situations and find novelty invigorating) are more likely to develop disorders such as drug addiction and attention deficit disorder (Eigsti et al., 2006). To explore underlying neurobiological differences that can give rise to high vs. low novelty reactivity, our lab uses Sprague Dawley rats that were selectively bred based on their novelty response: high novelty responders (HRs) and low novelty responders (LRs) (Stead et al., 2006). In previous studies, we found marked transcriptome changes in the brains of HR and LR rats during early postnatal development and adulthood (Cohen et al., 2017; McCoy et al., 2017, 2016). Looking across this body of work, a common molecular pathway that emerges as being different in the HR/LR brain includes numerous genes involved in synaptic transmission and cellular metabolism. Neurons rely on oxidative metabolism to provide high energy levels required for synaptic transmission and maintenance of resting membrane. Consequently, the primary aim of the present study was to characterize HR/LR differences in oxidative metabolism in multiple limbic brain regions that likely contribute to HR/LR emotional behavior differences.
A number of studies in humans suggest that the pathophysiology of several major psychiatric disorders involve altered brain metabolism and/or perturbed mitochondrial function (i.e., for reviews see (Marazziti et al., 2011),(Cuperfain et al., 2018)). For instance, human neuroimaging studies consistently report abnormal metabolic activity in limbic brain regions in depressed patients (Konarski et al., 2008; Lee et al., 2008; Videbech, 2000). Likewise, many patients suffering with mitochondrial diseases frequently exhibit psychiatric symptoms, including depression and suicidality (Boles et al., 2005; Inagaki et al., 1997; Marazziti et al., 2011), Such findings are supported by preclinical animal studies showing impaired metabolism in brains of rodents exposed to various types of chronic stress (de Vasconcellos et al., 2005; Madrigal et al., 2001; Rezin et al., 2008) , which are commonly used to model abnormal behaviors relevant to depression and anxiety. Another study used genetic knockout mice to perturb subunits of oxidative phosphorylation, alter ATP/ADP transport, or alter oxidative stress which disrupted the stress response, hippocampal gene expression and circulating metabolites (Picard et al., 2015). Furthermore, Harro and colleagues have performed multiple studies on oxidative metabolism across the brain to map neural activity within rodent models of stress and emotional disorders (Harro et al., 2014, 2011).
Our laboratory recently profiled the gene expression and levels of oxidative metabolism in the hippocampus, amygdala, and prefrontal cortex of HR/LR brains during early postnatal development (McCoy et al., 2016). We assessed metabolic activity in these brain regions by measuring the activity of cytochrome c oxidase (COX), the terminal enzyme of the electron transport chain leading to ATP production. Our study found decreased COX activity in the LR vs. HR hippocampus in the first postnatal week and decreased COX activity in LR vs. HR amygdala during the first two postnatal weeks. Correlational analyses of COX activity across developing limbic brains suggested that the limbic brain development timeline is desynchronized between HR/LR rats. In the current study, we hypothesized that HR/LR differences in expression of metabolism-related genes and COX activity would persist into adulthood, which could potentially contribute to HR/LR differences in limbic brain circuitry and adult behavior.
Section 2. Materials and Methods
Experiments were approved by the local University Committee on the use and care of the animals, and in accordance with the National Institutes of Health (USA, 2015) and the National Research Council (USA, 2011) guidelines on animal research.
2.1. Animals
Adult male HR and LR rats were obtained from the 7th generation of our in-house colony of bred HR/LR lines. These lines were derived as described in our previous work (Stead et al., 2006). Housing was maintained at 21-23°C and humidity at 50-55% on a 12:12 light-dark cycle (lights on/off at 6AM/6PM). Rats were housed in groups of 2-3 per cage with food and water ad libitum. Numbers of animals used in each experiment are noted within each subsection below.
2.2. Gene expression analysis
We previously published results of transcriptome studies that examined hippocampus and amygdala of early postnatal and adult male HR/LR rats (Cohen et al., 2015; McCoy et al., 2017, 2016). These prior analyses focused on comparisons of HR/LR control rats relative to other experimental groups (e.g., exposure to different maternal care conditions in one study (Cohen et al., 2015)). Since our current experiments were focused on metabolic differences in the adult HR/LR brain, we revisited the transcriptome data to examine expression of genes related to oxidative metabolism in control/unmanipulated adult male HR/LR hippocampus and amygdala (n=5 per HR/LR phenotype per brain region). These datasets were collected using NimbleGen Rat Gene Expression 12x135K Arrays (including 26,419 target genes with 5 probes/gene) can be found in the NCBI GEO repository (accession number: GSE88874) and initial procedural details are detailed in our previous publication (Cohen et al., 2015; McCoy et al., 2016).
The present data analysis included performing pathway enrichment analysis to identify functional pathways that may be altered due to disparate expression of various genes in HR/LR hippocampus and amygdala. We used ANOVA to contrast HR vs. LR samples within the a given region, restricting the criteria to genes that exhibit greater than 1.2 fold change (FC), p-value < 0.1; this yielded 1092 genes differing in the HR vs. LR hippocampus and 2286 genes differing in amygdala. While this is not conservative, we chose this strategy in order to maximize the possibility of discovering broad patterns within the larger dataset rather than focusing on individual genes. We performed enrichment analysis on the hippocampus and amygdala gene lists, and in doing so, applied statistical thresholds to identify functional pathways that were enriched within each dataset. Gene lists were imported into the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resource (ver. 6.8)(Huang et al., 2009a, 2009b). Through DAVID, we performed functional annotation clustering separately with the hippocampal and amygdalar gene lists. This tool allows one to reduce redundancy of similar annotations. We described the functional clusters of genes based on the top Up Keywords and/or KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment terms within the cluster. Finally, given our interesting in probing possible metabolic differences in the HR/LR brain, we focused a piece of our analysis on the functional group metabolic pathways to ascertain number of genes in these categories that were altered between HR/LR in the amygdala and hippocampus. These were selected from gene ontology lists and then genes meeting the criteria above (FC> 1.2, p< 0.1) were compared to these lists.
2.3. Oxygen Consumption in HR/LR hippocampal and amygdalar mitochondria
To isolate mitochondria for these experiments, adult male HR/LR rats were sacrificed at postnatal day 75 via rapid decapitation (n=6 per HR/LR phenotype). Brains were removed and the amygdala and hippocampus were gross dissected on ice and placed in ice cold isolation buffer (67 mM sucrose, 50 mM Tris-HCl, 50 mM KCl, 10 mM EDTA, 0.2% BSA). Mitochondria were isolated through differential centrifugation (Pandya et al., 2009; Sauerbeck et al., 2011) and then prepared for Seahorse XF Analyzer assays (Agilent, Santa Clara, CA). At the conclusion of this sample preparation process, two of the LR samples were deemed to be poor quality, so the sample sizes for Seahorse Assays was n=6/HR group and n=4/LR group.
During the the Seahorse XF analyzer assays, the oxygen consumption rate (OCR) is measured under the following conditions: (a) baseline/no stimulation; (b) 5 mM potassium (K) and adenosine diphosphate (ADP); (c) 4 μM Oligomycin (inhibits ATP synthase; ATP-linked OCR); (d) 4 μM Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP; depolarizes mitochondria;uncouples oxygen consumption from ATP synthesis to measure maximal respiration); and (e) 4 μM Antimycin A (inhibits complex III; non-mitochondrial OCR respiration). For experiment 1, the substrates applied were 5 mM pyruvate and 5 mM malic acid, which utilizes the entire electron transport chain. In experiment 2, the substrates applied were 20 mM succinic acid and 2 μM rotenone, this provides substrate for complex II and inhibits complex I, respectively. Assays were performed on four technical replicates. Values were normalized to total mitochondrial protein, which was measured through a standard bicinchoninic acid (BCA) assay. We calculated and compared basal respiration, mitochondrial respiration and reserve capacity in HR/LR samples. Reserve capacity was calculated as the percent change in OCR between FCCP treatment and basal respiration. Maximal respiration was the OCR after the FCCP treatment.
2.4a. In situ COX activity assay
This experiment was performed as previously described in Melendez-Ferro et al. (Melendez-Ferro et al., 2013). Brain tissue from adult (postnatal day 75) HR/LR rats was collected through rapid decapitation and flash frozen in isopentene on dry ice and stored at −80°C until processed (n=9 HR rats and n=7 LR). The brains were sectioned at 15 μm thickness in intervals of 300 μm then again stored at −80°C. On the day of histochemistry reaction, the brains were warmed to room temperature for 15 min and then reaction medium is applied and incubated for 30 min in the dark at 37°C. To terminate the enzymatic reaction, the sections were submerged in 4% paraformaldehyde at room temperature for 1 hr. Sections were washed in 0.1M sodium phosphate buffer, dehydrated and mounted using standard procedures. Along with the slides, we incubated a standard dot blot of known COX protein concentrations (0.03-2 μg) as described previously(Melendez-Ferro et al., 2013) to allow more accurate assessment of COX activity.
2.4b. Image analysis
Slides and dot blot were scanned on a MicroTek ScanMaker 9800XL in 16-bit grayscale settings without corrections. NIH ImageJ software (version 1.48v) was used to measure optical density (OD). An exponential standard curve were created from the OD values from the COX standards where R2=0.95. The images were then used to measure the activity of COX in select brain regions known to regulate emotional behavior: the hippocampus, amygdala, prefrontal cortex, and dorsal raphe. To define the anatomical structures, we referenced Paxinos & Watson brain atlas (6th edition). In the analysis of the hippocampus, we focused on the innermost molecular layer of the cornu ammonis (CA) region, which is where hippocampal neurons synapse, as well as upper and lower blades of the dentate gyrus (DG). In the amygdala, we examined 4 subnuclei that were reliably visible across experimental subjects: the lateral amygdala (LA), basolateral amygdala (BLA), basomedial amygdala (BMA), and central amygdala (CeA). We also analyzed COX activity in the prelimbic and infralimbic cortex, given the role of these cortical regions in regulating hippocampal and amygdalar function(Muir et al., 2019), and the dorsal raphe nucleus, a primary source of serotonin for the hippocampus and amygdala (Glover and Clinton, 2016). For each region, COX measurements were made in at least 4 sections per rat that were spaced 300 μm apart across the rostro-caudal extent of each brain region. Measurements were taken by at least two blinded, independent experimenters in ImageJ (Schneider et al., 2012), compared for consistency and averaged together for statistical analysis.
2.5. Correlation and post-hoc analysis of HR/LR COX activity
We carried out a secondary analysis with the HR/LR COX data using correlation matrices to determine inter-correlations of COX activity within certain limbic brain regions (i.e. within hippocampal subregions or within nuclei of the amygdala) as well as correlations across regions (i.e. hippocampus or amygdala with prefrontal cortex or dorsal raphe). R studio (Version 1.0.153, R. RStudio, Inc., Boston, MA) was used to calculate and display the correlations in a color map. The rcorr() function downloaded from the “Hmisc” package was used to calculate the correlation coefficients and p-values. These data were then visualized by way of a color map using the corrplot() function from the “corrplot” package.
Principle component analysis (PCA) is a statistical tool used to reduce the dimensions of a dataset to find inter-correlations between many variables. Data are reduced in such a way that the components of variance are apportioned to 9 principle component dimensions. As the first two component capture 69% of the variability within the dataset. Therefore, we presented these using a biplot scatterplot that depicts distinct brain regions with eigenvectors and principal component 1 (PC1) and principal component 2 (PC2) on the axes. We created a PCA biplot using the ggbiplot() function from the “ggbiplot” package in R studio (Version 1.0.153, R. RStudio, Inc., Boston, MA).
2.6. Statistical Analysis
Data were analyzed using GraphPad Prism Software (Version 6.0 for Windows, GraphPad Software, La Jolla, California USA, www.graphpad.com) Prior to any analysis, all datasets were verified to be normally distributed using the D’Agostino & Pearson omnibus normality test. If the data were not normally distributed, Mann-Whitney U ranks test was applied. For the COX data, variances for each experimental group were compared with F test; if variances did not differ between groups, Student t-tests were used to compare metabolic measures in HR vs. LR rats within each brain region. If the variances were found to be significantly different, Welch’s corrected T-test was used. For the oxygen consumption rate measurements, 2-way ANOVAs were performed, with independent variables being HR/LR phenotype and assay condition (which changed across the Seahorse assay to assess different components of electron transport chain function); Tukey’s post-hoc tests were used as needed.
Section 3. Results
3.1. Metabolic gene expression differences in HR/LR hippocampus and amygdala
Our previous transcriptome study found distinct HR/LR gene expression patterns in the early postnatal hippocampus and amygdala as well as decreased COX activity at postnatal day 7 in CA regions of the hippocampus and amygdala (McCoy et al., 2016). The present analysis used our previously collected transcriptome data (Cohen et al., 2015; McCoy et al., 2017) to determine whether HR/LR gene expression changes in amygdala and hippocampus persist into adulthood. We began by performing pathway cluster analysis in DAVID, a tool that clusters differentially expressed genes based on biological processes that the genes are known to be involved in. Table 1 displays the top 10 gene clusters named according to the top Up Keyword Enrichment term and/or KEGG pathway for each brain region. In the hippocampus, we found that ion channel-related genes are highly enriched in the differentially expressed gene list (including those in Cluster 5, 8 and 10; top of Table 1), which may reflect HR/LR changes in hippocampal cell excitability. In the amygdala, major pathways that differed between HR/LR animals related to synaptic transmission and components of the cell membrane (including Cluster 1,6, 8, and 9; bottom of Table 1).
Table 1. Cluster analysis of HR/LR hippocampus and amygdala transcriptome data points to alterations of metabolism and mitochondrial function.
Previous gene expression microarray datasets from adult male HR/LR amygdala and hippocampus were used for DAVID cluster analysis. This analysis identified functional clusters among genes that were differentially expressed in the HR/LR brain. The top three functional cluster terms are shown along with the total number of genes in that cluster, number of genes altered between HR and LR, minimum percentage of enriched genes within one term in the cluster, and the maximum p-value of individual pathways within the cluster. Overall cluster enrichment score is listed for each cluster. The top 10 clusters are ranked from highest to lowest enrichment score.
| DAVID cluster analysis of HR/LR adult hippocampal gene expression | |||||
|---|---|---|---|---|---|
| Cluster # | Up Keyword Enrichment Term | # of altered in pathway | Minimum % of enriched genes | Cluster Enrichment Score | Maximum P-value of Pathways |
| 1 | Cell membrane, Membrane, Receptor | 174 | 17.791 | 8.468 | 2.30E-05 |
| 2 | Cell membrane, Receptor, Transducer | 83 | 13.599 | 6.109 | 2.58E-07 |
| 3 | Cell junction, Synapse, Postsynaptic cell membrane | 22 | 2.249 | 5.750 | 5.76E-06 |
| 4 | Lipoprotein, Palmitate | 29 | 2.965 | 4.765 | 1.27E-06 |
| 5 | Sodium transport, Sodium, Sodium channel | 5 | 1.534 | 3.033 | 0.039 |
| 6 | Mitochondrion, Transit peptide | 31 | 3.170 | 2.946 | 2.53E-04 |
| 7 | Glycoprotein, Disulfite bond, Secreted | 73 | 7.464 | 2.746 | 5.62E-04 |
| 8 | Ion channel, Voltage-gated channel | 16 | 1.636 | 2.306 | 9.87E-04 |
| 9 | Glucose metabolism, Carbohydrate metabolism | 5 | 0.511 | 1.900 | 9.43E-03 |
| 10 | Potassium transport, Potassium, Potassium channel | 9 | 0.920 | 1.885 | 1.34E-02 |
| DAVID cluster analysis of HR/LR adult amygdalar gene expression | |||||
| Cluster # | Up Keyword or KEGG Pathway Enrichment Term | # of altered in pathway | Minimum % of enriched genes | Cluster Enrichment Score | Maximum P-value of Pathways |
| 1 | Cell junction, synapse, postsynaptic cell membrane | 54 | 2.410 | 25.914 | 6.17E-16 |
| 2 | Mitochondrion, Mitochondrion inner membrane | 60 | 2.679 | 16.028 | 4.71E-16 |
| 3 | Mitochondrion, Transit peptide | 83 | 3.705 | 12.492 | 4.75E-14 |
| 4 | Nucleotide-binding, Kinase, ATP-binding | 139 | 6.205 | 8.776 | 3.60E-18 |
| 5 | Long-term potentiation, Circadian entrainment, Amphetamine addiction | 25 | 1.116 | 6.753 | 4.83E-06 |
| 6 | Glutamergic synapse, Circadian entrainment, Retrograde endocannabinoid signaling | 33 | 1.473 | 6.228 | 1.45E-05 |
| 7 | Ribonucleoprotein, Ribosome, Ribosomal protein | 60 | 2.679 | 6.028 | 6.65E-11 |
| 8 | GABAergic synapse, Retrograde endocannabinoid signaling, Morphine addiction | 30 | 1.339 | 5.544 | 2.67E-05 |
| 9 | Membrane, Transmembrane, Transmembrane helix | 689 | 30.759 | 5.501 | 1.19E-01 |
| 10 | Glycoprotein, Disulfide bond | 303 | 13.527 | 5.008 | 1.87E-06 |
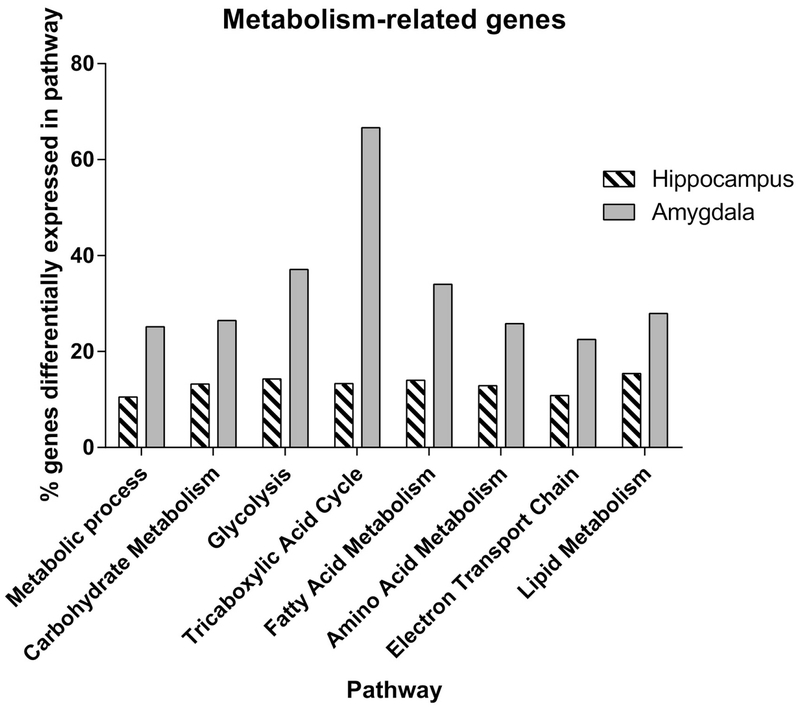
The enrichment analysis also pointed to marked HR/LR differences in genes involved in mitochondrial function and other cellular metabolism processes in both the hippocampus and amygdala. These findings led us to classify the gene expression data based on specific metabolic pathways that genes were involved in (e.g., genes involved in carbohydrate metabolism, glycolysis, fatty acid metabolism, electron transport chain, etc.). Figure 1 shows the percentage of genes within each of these pathways that were altered in the adult HR vs. LR hippocampus and amygdala. In hippocampus, approximately 10% of the genes within each of the different metabolic pathways were altered between HR/LR animals. In amygdala, an average of 20% of the genes within each pathway (up to 40-60% of genes in select pathways) were altered between HR/LR, with the tricarboxylic acid cycle being most affected.
Figure 1. Myriad genes involved in mitochondrial function and cellular metabolism are differentially expressed in the adult HR/LR hippocampus and amygdala.
Percentage of genes within several distinct metabolic pathways that were found to be differentially expressed in adult HR/LR hippocampus and amygdala. Genes were considered differentially expressed between HR/LR groups based on a fold change of ±1.2 or greater and a p-value < 0.1 with BH correction.
3.2. Mitochondrial respiration in HR/LR hippocampus and amygdala
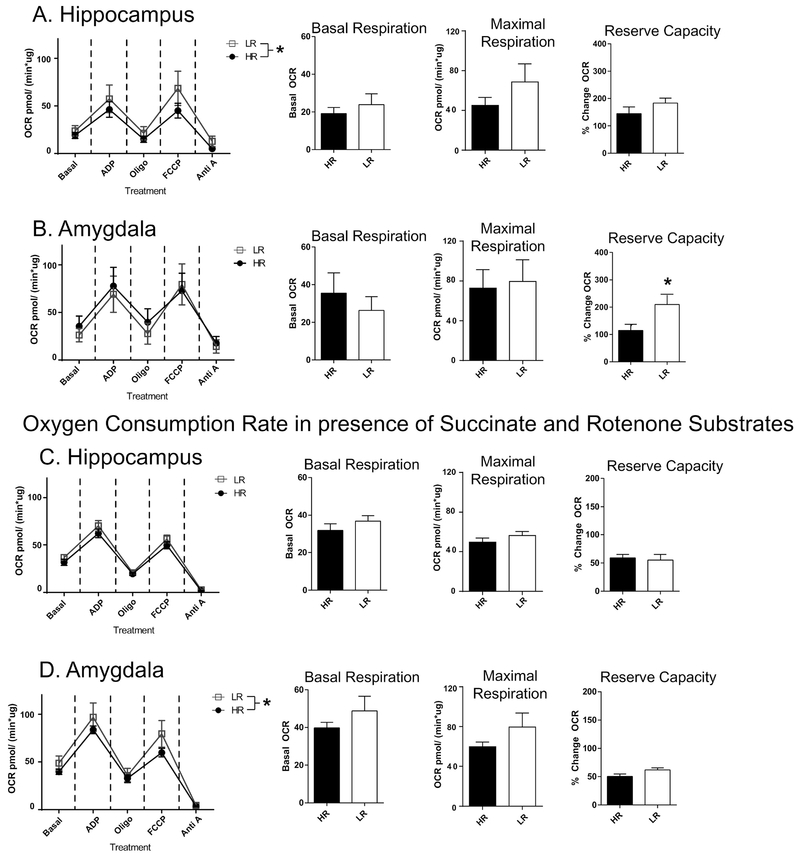
Based on our observation of marked HR/LR gene expression differences within numerous genes involved in mitochondrial function and other aspects of cellular metabolism, we hypothesized that oxygen consumption rate of isolated mitochondria and/or in COX activity may also differ in brain tissue from HR/LR rats. We began by performing the oxygen consumption assays in isolated mitochondria from HR/LR whole hippocampus and amygdala. First, our assay results indicated that mitochondria isolates from HR/LR tissue were responsive and had minimal leak. Our first Seahorse assay (experiment 1) performed the mitochondrial oxygen consumption challenge assay in the presence of pyruvate and malate to test whether the citric acid cycle or electron transport chain exhibited HR/LR differences. In isolated hippocampal mitochondria, we found an overall effect of HR/LR phenotype across conditions of the oxygen consumption assay (F(1,40)=4.525, p=0.0396), but no phenotype x assay condition interaction. In this part of the experiment, there were no effects of HR/LR phenotype on baseline respiration, maximal respiration, or reserve capacity (Fig. 2A). In amygdalar mitochondria, there was no overall effect of HR/LR phenotype across the individual assay conditions, and no significant phenotype differences in baseline or maximal respiration. There was, however, significantly increased reserve capacity in LR vs. HR amygdala (t(8)=2.433, p= 0.0410; Fig. 2B).
Figure 2. Oxygen Consumption Rate (OCR) in isolated mitochondria of HR/LR hippocampus and amygdala.
(A-B) The first oxygen consumption assay was conducted in the presence of pyruvate and malate to assess function of the full electron transport chain in either hippocampal (A) or amygdala (B) tissue. Line graphs at left depict OCR at baseline and under a series of assay conditions, including application of: adenosine diphosphate (ADP); Oligomycin (Oligo); carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP); or Antimycin A (Anti A). Bar graphs at right display levels of basal respiration; maximal respiration (OCR after FCCP treatments); and reserve capacity (OCR between FCCP treatment and basal respiration). (C-D) The second assay assessed hippocampal (C) and amygdalar (D) OCR in the presence of succinate and rotenone to evaluate mitochondrial oxidative metabolism with complex I uncoupled. * indicates p-value < 0.05.
In amygdalar mitochondria, there was no overall effect of HR/LR phenotype across the individual assay conditions; there were no significant phenotype differences in baseline or maximal respiration, although there was significantly increased reserve capacity in LR versus HR amygdala (t(8)=2.433, p= 0.0410; Fig. 2B). The oxygen consumption assay in experiment 2 (Fig. 2C and D) was performed in the presence of succinate, the substrate for complex II, and rotenone to inhibit complex I. In this case, there were no significant differences in HR/LR hippocampus in the various measures of oxygen consumption rate. In amygdala, there was a main effect of HR/LR phenotype across assay conditions (F(1,40)=4.913, p=0.0324), but no phenotype x condition interaction. We found no group differences in basal respiration, maximal or reserve capacity in HR/LR amygdala when complex I is inhibited.
3.3. COX activity in the adult male HR/LR limbic brain
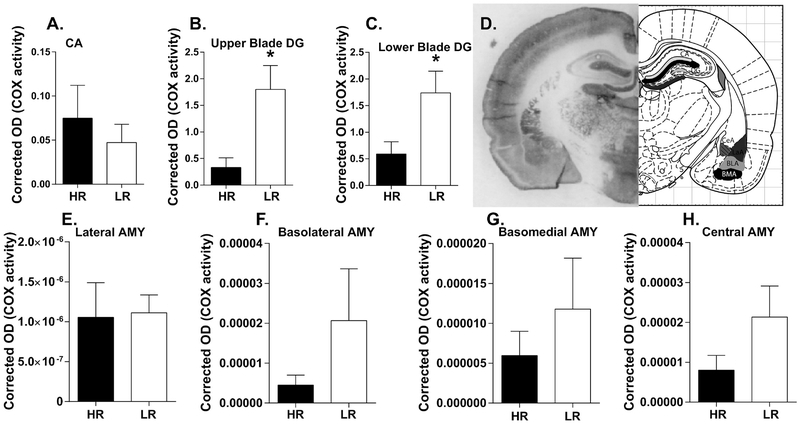
Our next study used a COX histochemical activity assay to ascertain potential HR/LR metabolic differences within select brain regions that contribute to emotional behavior regulation. The first phase of COX analysis focused on the hippocampus and amygdala as in the gene expression and mitochondrial studies. In the hippocampus, there were no HR/LR differences in COX activity in the CA region (Fig. 3A). In the DG, LR rats displayed increased COX activity compared to HRs in both the upper blade of the DG (t(7.904)=3.022, p=0.0167; Fig. 3B) and lower blade of the DG (t(14)=2.602, p=0.0209; Fig. 3C). We found no significant HR/LR differences in COX activity within any of the amygdalar nuclei examined (Fig. 3E–H).
Figure 3. Cytochrome C oxidase (COX) activity in adult HR/LR hippocampus and amygdala.
(A-C) COX activity was assessed in the innermost molecular layer of the cornu ammonis (CA) region, where many hippocampal neurons form synapses (A), as well as in the upper blade (B) and lower blade (C) of dentate gyrus (DG). (D) Representative image of brain tissue processed for the in situ COX assay at the level of rostral hippocampus and amygdala (bregma −2.04 mm) Regions of interest are shown in black and grey on corresponding atlas images, which were adapted from “The Rat Brain” (6th Edition) by Paxinos & Watson). (E-H) COX activity was also measured in four nuclei of the amygdala: the lateral amygdala (LA; E); basolateral amygdala (BLA; F); basomedial amygdala (BMA; G); and Central amygdala (CeA; H). * indicates p-value < 0.05.
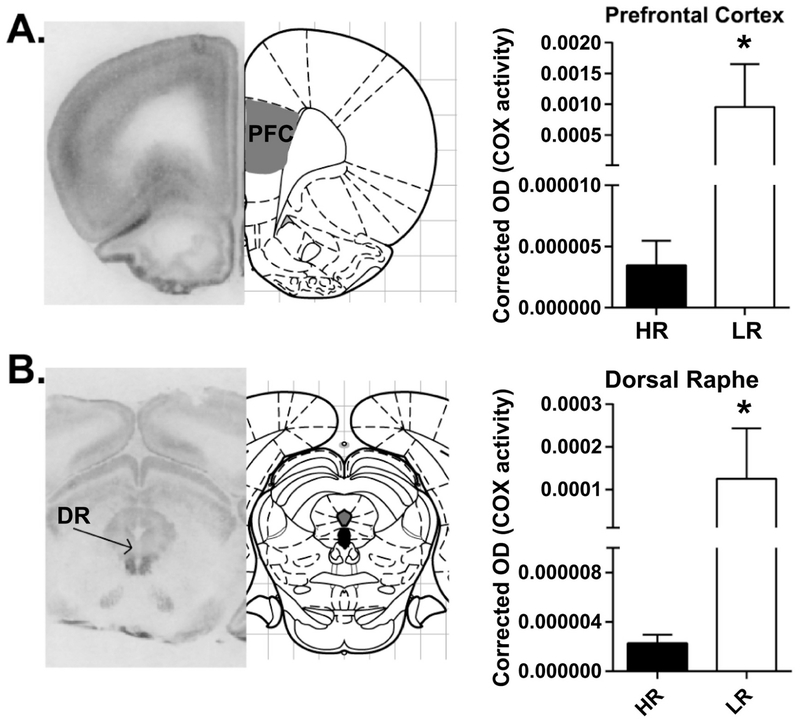
To broaden our COX analysis in adult HR/LR rats, we expanded our studies to ascertain COX activity in three additional brain regions known to participate in emotion regulation – the prelimbic cortex, infralimbic cortex, and dorsal raphe. Here we found increased COX activity in the prelimbic cortex of LR vs. HR rats (U=4, Sum of ranks: HR=19, LR=59, p=0.0303: Fig. 4A), but no HR/LR difference in the infralimbic cortex (HR: 2.38E-6 ± 1.26E-5, LR: 8.94E-5 ± 4.82E-5, t(5.007)=1.803, p=0.1312). In the dorsal raphe, we found increased COX activity in LRs vs. HRs (U=3, Sum of ranks: HR=24, LR=42, p=0.0303: Fig. 4B).
Figure 4. COX activity in prelimbic cortex, infralimbic cortex, and dorsal raphe of HR/LR adults.
Left panels show representative images of brain sections processed for the in situ COX assay at the level of the prelimbic and infralimbic cortices (bregma +2.76) or dorsal part of dorsal raphe (DR; bregma −7.92 mm) alongside adapted atlas images from Paxinos & Watson “The Rat Brain” (6th Edition). Right panels show relative COX activity in HR/LR prelimbic cortex and DR. * indicates p-value < 0.05.
3.4. Correlational analysis of COX activity across limbic brain regions
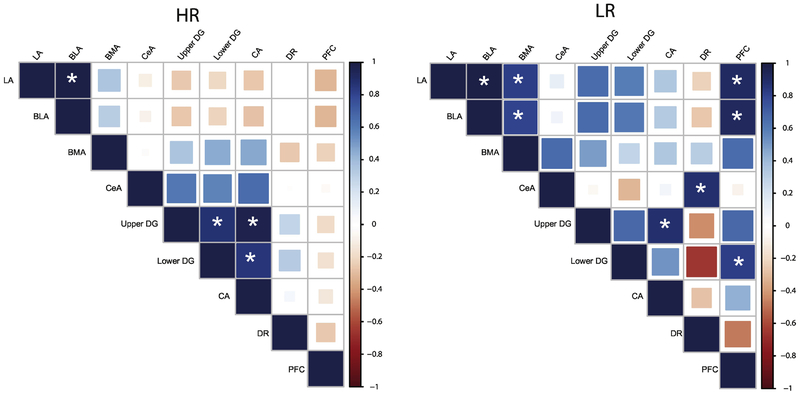
In order to extend our examination of COX activity in select limbic brain regions of adult HR/LR rats, we utilized a correlation matrix analysis (described in one of our earlier publications (McCoy et al., 2016)) to assess activity across brain regions. Color maps in Fig. 5 depict COX activity correlations among the several regions that were examined in adult HR/LR brains. We first looked at inter-region correlations for (a) the amygdala; and (b) the hippocampus. Within the amygdala, there were positive inter-nuclei correlations among 3 of the 4 amygdalar nuclei examined (LA, BLA, BMA), but less correlation of activity with CeA; interestingly, these effects were more prominent in LR vs. HR animals. In the hippocampus, there were also positive inter-region correlations among the CA and blades of the DG; in this instance, the effect was stronger in HRs than in LRs.
Figure 5. Inter-correlations of COX activity within several limbic brain regions in adult HR/LR rats.
Correlation matrices were used to determine Inter-correlations of COX activity within certain regions (i.e. within subregions of the hippocampus or the amygdala) as well between regions. The color and size of each tile reflects the direction and strength of the correlation respectively, as indicated by the color bar on the right. The asterisk indicates a statistically significant correlation (P<0.05). Separate correlation matrices are shown for HR (left) and LR (right). The color of squares at each convergence of a row and column indicates the level of correlation according to the color bar on the right. Inter-correlations within the amygdala were positive, particularly in LR animals. Within the hippocampus, there were positive interregional correlations, although the effect was more prominent in HRs. HR/LR rats exhibited distinct cross-regional correlations (i.e. between hippocampus and the prefrontal cortex or dorsal raphe). Abbreviations: lateral amygdala (LA); basolateral amygdala (BLA); basomedial amygdala (BMA); central amygdala (CeA); DG, dentate gyrus; CA, cornu ammonis; dorsal raphe (DR); Prl, prelimbic cortex.
Examining correlations of COX activity across the different brain regions revealed several interesting HR/LR phenotypic differences. For instance, HRs showed a negative correlation of COX activity between hippocampal regions and LA/BLA nuclei of the amygdala and moderate positive correlations between hippocampal regions and BMA/CeA nuclei. LRs, on the other hand, showed moderate to strong positive correlations between hippocampal COX activity and that in LA, BLA, and BMA amygdala, but no correlation with CeA. There was little to no correlation between COX activity in the dorsal raphe and amygdala of HRs, but a weak to moderate correlation of these measures in LRs. There was a weak positive correlation between COX activity in the dorsal raphe and hippocampus of HRs, but a strong negative correlation of these measures in LR. Lastly, for the prelimbic cortex, there were little to no correlations of COX activity in the prelimbic cortex and other regions examined in HRs; however, in LRs, there was a strong correlation between COX activity in the prelimbic cortex, amygdala, and hippocampus (Fig. 5)
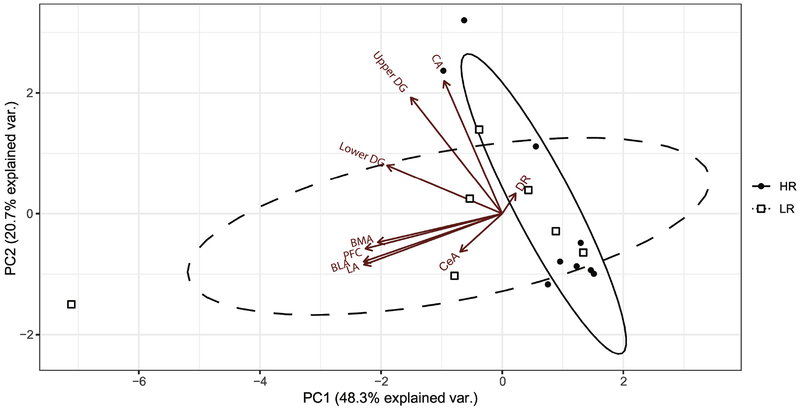
For our final phase of COX activity analysis, we used PCA to further illustrate relationships between COX activity levels across regions of the HR vs. LR adult brain. Eigenvector loadings based on the variation of COX activity within each brain region were computed to determine vector inter-correlations as well as correlations with the top two components that together contributed to 69% of the variability in our data – principle component (PC)1 and PC2. In Fig. 6, the following brain regions all had large negative loadings on PC1: LA, BMA, BLA, prelimbic cortex, upper blade of DG, and lower blade of DG. A negative loading with PC1 indicated a negative correlation with PC1. Therefore, data points with higher PC1 values would have lower COX activity for the aforementioned regions. COX activity within three amygdalar nuclei (LA, BMA, and BLA) was strongly correlated with each other as well as with COX activity in the prelimbic cortex. All three hippocampal regions (CA hippocampus, upper and lower blades of the DG) were inter-correlated with each other and had negative correlations with PC1 and PC2. CA hippocampus, however, was more negatively correlated with PC2 than with PC1. PC2 accounted for 22.6% of variability within the dataset and was predominantly influenced by COX activity in the DR, CeA, and CA hippocampus. COX activity in the CA hippocampus was the only brain region examined that negatively correlated with PC2.
Figure 6. Principle Component Analysis (PCA) Biplot of COX data in adult HR/LR limbic brain.
PCA was used to examine inter-correlations between COX activity measures across the amygdala, hippocampus, prefrontal cortex, and dorsal raphe or HR/LR rats by reducing the data into two major components of variance – Principle Components (PC) 1 and 2. PC1 (shown on x-axis) explains 48.3% of the variation observed in COX activity within HR/LR brain regions. PC2 (shown on y-axis) explains 20.7% of the variation in COX activity within HR/LR brain regions. Arrows labelled with different brain regions represent vectors that indicate direction and magnitude of variation of COX data for each brain region. The solid black line ellipse represents the range of HR samples (individual animals shown in small black-filled circles). The dashed-line ellipse represents the range of the LR samples (individual animals shown in small open squares). The distinct shape and spread of the HR vs. LR ellipses illustrates the COX differences and differential regional inter-relatedness between phenotypes. Abbreviations: lateral amygdala (LA); basolateral amygdala (BLA); basomedial amygdala (BMA); central amygdala (CeA); DG, dentate gyrus; CA, cornu ammonis; dorsal raphe (DR); Prl, prelimbic cortex. See supplementary material for eigenvector loadings on PC1 and PC2.
Section 4. Discussion
Many studies report altered brain metabolism and/or mitochondrial dysfunction in patients suffering with major psychiatric disorders as well as in animal models relevant to human emotional disorders (Cuperfain et al., 2018; Marazziti et al., 2011). Prior work with the HR/LR rat model demonstrated that the low novelty reactive LR rats display high levels of anxiety- and depression-like behavior relative to their HR counterparts (Flagel et al., 2014). The distinct HR/LR behavioral phenotypes are likely driven by a number of neurobiological differences, although much work to-date points to particular differences in the hippocampus and amygdala (Clinton et al., 2011b; Cohen et al., 2015; Simmons et al., 2012; Turner et al., 2011). Dramatic gene expression differences occur in the early postnatal as well as adult HR vs. LR amygdala and hippocampus (Cohen et al., 2017; McCoy et al., 2017, 2016), and a major functional pathway found to differ in the HR vs. LR brain pertains to genes involved in myriad metabolic processes. Thus, the primary goal of the present study was to more fully investigate metabolic changes in the adult HR vs. LR brain by a) revisiting prior transcriptome data for additional in-depth analysis; b) examining mitochondrial function in HR/LR brain tissue; and c) examining COX activity, an indicator of metabolic function, in the adult HR/LR hippocampus, amygdala, and other limbic regions (prelimbic cortex, infralimbic cortex, and dorsal raphe).
4.1. HR/LR rats have distinct brain metabolic profiles
Previous transcriptome studies revealed myriad gene expression changes in the early postnatal HR vs. LR hippocampus and amygdala, with pronounced changes in genes involved in metabolic pathways (Clinton et al., 2011b; McCoy et al., 2016). Our present analyses of adult HR/LR amygdala and hippocampus transcriptome data show that these metabolism differences persist into adulthood, with numerous genes broadly related to metabolism being differentially expressed in adult HR/LR hippocampus and amygdala. When the list of altered genes classified based on specific types of metabolism (e.g., carbohydrate metabolism, glycolysis, fatty acid metabolism, electron transport chain), we found that HR/LR differences exist across multiple types of metabolic processes. Although these data point to altered metabolism in both the HR vs. LR hippocampus and amygdala, these phenotypic differences were more pronounced in the amygdala.
In order to build on the observed HR/LR metabolism-related gene expression differences, we chose two assays to examine metabolic function in the HR vs. LR brain: (1) a mitochondrial assay to measure oxygen consumption under several biochemical conditions, and (2) an in situ COX activity assay to provide an indication of metabolic activity in select brain regions. Results of the mitochondrial assay indicated that (a) mitochondria were viable and healthy in both HR and LR brain tissue; and (b) there were no phenotypic differences in baseline respiration. We found increased reserve capacity during respiration in oxidative metabolism in the amygdala of LR rats only when complex I is active. Within the hippocampus, experiment 1 revealed that LRs have increase oxygen consumption rate through all treatments. This did not occur in experiment 2, which may be due to the inhibition of complex I (thereby reducing the proton gradient across the membrane) in experiment 2 conditions that prohibits a greater reserve capacity to be generated. Neurons are thought to utilize up to 80% of their maximum respiration when firing therefore this change could point to a functional change in synaptic transmission (Fried et al., 2014). Such possibilities will be pursued in future studies.
Our COX histochemistry experiment extended our HR/LR metabolism analyses beyond the hippocampus and amygdala to also include additional limbic brain regions – the prelimbic cortex, infralimbic cortex, and dorsal raphe – which play key roles in regulating emotional behavior, in part, through interconnections with the hippocampus and amygdala (Glover and Clinton, 2016; Muir et al., 2019). We found increased COX activity in the dentate gyrus of LR vs. HR rats, but no phenotypic difference in the CA region of the hippocampus and no difference in any of the four amygdalar nuclei examined. We also found increased COX activity in the prelimbic cortex and dorsal raphe of LR vs. HR rats, with no phenotypic difference in the infralimbic cortex.
The present study is the first to examine COX activity in the adult HR vs. LR brain. COX activity is typically viewed as an indicator the baseline neural activity (Arias et al., 2015; Hescham et al., 2014). Another approach frequently used to examine neuronal activation patterns in various rodent models (including HR/LR rats) is to measure expression of the immediate early gene cfos. Numerous studies have examined cfos expression throughout the brain to identify brain circuits that are activated in response to a wide variety of stimuli, including exposure to stress or following learning (Dragunow and Faull, 1989; Kaczmarek, 1995; Robertson, 1992). Our prior work with HR/LR rats found that exposure to various stressors elicited both distinct behavioral responses as well as disparate cfos neural activation patterns in HR vs. LR animals. For example, when adult male HR/LR rats were subjected to the Resident Intruder paradigm where a novel male rat was introduced into the homecage of and HR or LR resident, HR rats displayed high levels of aggression while LRs did not; subsequent cfos studies in HR/LR brain tissue showed that homecage intrusion stress elicited higher cfos expression in the dorsal raphe of LR vs. HR rats (Kerman et al., 2011) but reduced activation in the hippocampus and other forebrain regions (Clinton et al., 2011a). Another study examined behavior and cfos activation in HR/LR rats subjected to the defensive burying task, a test of stress coping style where rats are exposed to an electric probe to determine if they choose an active coping style (burying the shock probe) or a reactive/passive coping style (freezing in the corner of the cage and thereby avoiding the probe). In this test, LR rats showed high levels of reactive stress coping while HRs were high ‘active copers’; results of the cfos study showed that LRs (compared to HRs) exhibited increased cfos in the amygdala after exposure to shock (Cohen et al., 2017). Our current COX data together with these earlier cfos findings suggest that HR/LR rats exhibit both baseline differences in metabolic activity (indicated by COX) as well differences in acute activation of brain circuits in response to stressful stimuli (indicated by cfos).
An interesting feature of the present COX findings in HR/LR adult brain is that the direction of phenotype differences stand in contrast to our earlier COX results in developing HR/LR animals. That earlier work revealed decreased COX activity in the amygdala, CA hippocampus, and dentate gyrus of LR vs. animals at postnatal day (P) 7, with fewer (if any) differences at later ages P14 and P21 (McCoy et al., 2016). These distinct patterns of HR/LR COX activity differences in early postnatal life vs. adulthood are not particularly surprising. The brain undergoes dramatic transformation during early life through adolescence and adulthood. Our work in early postnatal HR/LR rats suggest that they exhibit distinct neurodevelopmental trajectories, which could plausibly lead to their characteristic behavioral and neural differences in adulthood. It seems possible that disparate formation of HR vs. LR brain circuits as well as distinct life experiences, such as LRs’ lifelong experience of high anxiety and different hypothalamic pituitary adrenal (HPA) stress axis reactivity (Clinton et al., 2008, 2011b; Kerman et al., 2012), could result in different levels of metabolic activity (COX activity) in limbic brain.
Correlational analyses of the current COX activity data suggests that HR/LR animals exhibit distinct intra-hippocampal and intra-amygdalar connectivity relative to one another, in addition to differences in connectivity across brain regions examined. For instance, LRs (compared to HRs) showed stronger correlations between prelimbic cortex COX activity and that in hippocampus and amygdala as well as stronger correlation between dorsal raphe COX activity and hippocampus. Additionally, through PCA analysis, we found that regardless of behavioral phenotype, the prelimbic cortex eigenvector was more correlated with those of the amygdalar nuclei than that of the hippocampus. Interestingly, the dorsal raphe vector was the least correlated brain region in the PCA analysis. It would be interesting to interrogate potential HR/LR neural circuit differences using tract tracing or neuroimaging approaches to determine whether they do in fact showed greater or lesser connectivity between these different limbic brain regions.
4.2. Metabolic differences in other animal models relevant to emotional disorders
Several previous studies have examined brain metabolism in other animal models relevant to anxiety and depression, including a substantial body of work by Harro and colleagues (Harro et al., 2014, 2011; Kanarik et al., 2011, 2008; Kanarik and Harro, 2018; Mallo et al., 2009; Matrov et al., 2019, 2007). As an example, one study by Harro et al examined COX activity across several brain regions within five rodent models relevant to depression, including animals exposed to maternal deprivation, chronic variable stress, or chronic social defeat (Harro et al., 2014). One of their overall findings was a general increase in COX activity across several brain regions of ‘stress vulnerable’ animals known to exhibit high levels of depression- and anxiety-like behavior. Likewise, other selectively-bred rat lines known to display high levels of anxiety- and/or depression-related behavior (akin to our LR rats) exhibit brain metabolism alterations (typically increased metabolism in several limbic regions relative to ‘normal’/non-anxious/depressive-like rats), including the High Anxiety Bred (HAB) rats bred for high anxiety in the Elevated Plus Maze (Filiou et al., 2014, 2011), the Naples Low Excitability (NLE) rats (Gonzalez-Lima and Sadile, 2000); Wistar rats bred for Low Exploratory activity (Matrov et al., 2007), and a congenitally helpless rat model (Shumake et al., 2002, 2001; Shumake and Gonzalez-Lima, 2003). Thus, our present findings of increased COX activity in multiple brain regions of adult LR (vs. HR) rats are broadly consistent with results from these other models of emotional dysfunction.
A common feature of animal models relevant to emotional disorders (including the HR/LR model and others noted above) is that animals that exhibit high levels of anxiety- and depression-like behavior also often exhibit HPA stress axis abnormalities (Clinton et al., 2014; Kerman et al., 2011). There has been speculation that chronic stress and/or dysregulated HPA stress axis function may lead to altered brain metabolism. For instance, work in adult Wistar rats exposed to early life maternal separation showed reduced complex II-III activity in hippocampus, amygdala and prefrontal cortex (Della et al., 2013). Another study found that adrenalectomized females exposed to cortisol for one hour before tissue collection showed increased mitochondrial respiration in whole cerebrum (Roosevelt et al., 1973). Chronic variable stress exposure lead to reduced oxidative activity in the hippocampus, amygdala and other regions, although, interestingly, this effect was reversed when the chronically stressed rats had their serotonin system lesioned (Kanarik et al., 2008). Chronic social defeat has also been shown to broadly suppress COX activity across multiple limbic regions, including amygdala and hippocampus (Kanarik et al., 2011). It is interesting to note that several of these chronic stress models elicit reduced COX activity in several adult limbic brain regions, in contrast to the genetic/selectively-bred models, including LRs and others noted above and as well in a previous cross analysis of these two types of animal models (Harro et al., 2014). Additionally, as mentioned in the Introduction, neuroimaging studies in depressed patients have reported diminished metabolic activity in select limbic brain regions (Konarski et al., 2008; Lee et al., 2008; Videbech, 2000). Taken together, findings from clinical studies as well as those from preclinical animal models illustrate ways that inborn biological factors and environmental factors (such as stress) influence brain metabolism and emotion.
Interestingly, recent studies have connected hippocampal neurogenesis and mitochondrial function (Beckervordersandforth et al., 2017; Knobloch et al., 2017), so a potentially important consideration to keep in mind with regard to our present findings is whether HR/LR metabolic differences (particularly in the hippocampus) relate to possible cell proliferation differences. Previous studies in early postnatal HR/LR rats found that LRs have higher rates of proliferation in the dentate gyrus compared to HRs during the first weeks of life (Clinton et al., 2011b). Adult LR rats also exhibited lower levels of adult neurogenesis relative to HRs, and this deficit could be rescued by treatment of the neurotrophic factor FGF2 (Perez et al., 2009). Other work showed that adult or adolescent exposure to cocaine differentially impacted hippocampal cell proliferation in HR and LR animals, which is interesting considering the marked behavioral differences that HR/LR rats show in response to cocaine and other drugs of abuse (Garcia-Fuster et al., 2017, 2010). Future studies will address the connections between metabolic function and neurogenesis and proliferation in HR/LR rats at baseline and following different environmental manipulations.
4.3. Limitations and Future Directions
A limitation of the present study is that it examined gene expression, mitochondrial function, and COX activity only in male HR/LR rats. Our previous work has found that HR/LR females display a number of similar behavioral differences that were initially observed in males, with LR females showing generally higher levels anxiety- and depression-like behaviors, as well as diminished reactivity to novelty and decreased response to psychostimulants relative to HR females (Cummings et al., 2011; Davis et al., 2008; Stead et al., 2006). A limited number of prior studies of COX activity in other animal models of emotional dysfunction included females and suggest possible sex differences. For instance, Mällo et al found that chronic stress lead to reduced COX activity in male but not female animals with a lower positive affect (measured through less 50-kHz ultrasonic vocalizations) in many brain regions, including raphe and amygdala. They also observed overall lower COX activity in females than males (Mallo et al., 2009). Spivey et al reported that maternal separation stress lead to altered COX activity levels in offspring brains, but found different effects in males vs. females (Spivey et al., 2011). Maternal separation exposure lead to diminished COX activity in several brain regions of females, including dorsolateral, orbital frontal, posterior parietal, and cerebellar cortical regions. In contrast, maternal separation lead to increased COX activity in the parietal cortex of male offspring (Mendez-Lopez et al., 2009).
Another limitation of the present work is that we focused most of our analyses on a circumscribed set of brain regions. Our gene expression and mitochondrial studies focused on the hippocampus and amygdala only, and the COX experiment included these plus three others – the prelimbic cortex, infralimbic cortex, and dorsal raphe. It is possible that HR/LR rats exhibit metabolic differences in other regions important for emotional regulation. Some of our prior work identified immediate early gene expression (cfos) differences in regions such as the lateral septum and subnuclei of the hypothalamus following exposure to different types of emotional stressors, so it may be useful for future COX experiments to extend to include these and other regions to obtain a more complete picture of HR/LR brain metabolism differences. Moreover, a general limitation of the COX assay is that it permits examination of metabolic activity levels at a region-wide but not cell-specific level. Brain regions like the hippocampus and amygdala are comprised of numerous cell types that may each exhibit distinct metabolic characteristics. Future studies must be done to gain a deeper understanding of whether HR/LR metabolic changes occur within specific cell populations and how those changes could contribute to their disparate phenotypes.
Section 4.4. Conclusion
Results of our present studies coupled with prior work in other animal models relevant to emotional disorders illustrate that metabolic dysfunction, particularly oxidative metabolism, occurs in brains of individuals prone to display high levels of anxiety- and depressive-like behavior. These preclinical findings are broadly consistent with observations of metabolic dysfunction in brains of psychiatric patients. Model animals such as the bred HR/LR rats offer a tool that can be exploited to better understand the relationship between brain metabolism and emotional behavior. Ultimately, such work in preclinical models may shed light on neurobiological mechanisms at play in psychiatric disorders, which can pave the way towards improved treatment strategies.
Supplementary Material
Highlights.
Mitochondrial gene expression is altered in adult anxiety-prone LR rat brain.
LRs have higher mitochondrial oxygen consumption in hippocampus and amygdala.
Higher COX activity observed in LR vs. HR hippocampus, cortex, and raphe.
Correlation analysis suggests distinct connectivity in LR vs. HR brains.
Acknowledgements
This work was supported by the National Institutes of Health (NIH) R01MH105447-01 (SMC). We would like to thank Adele Addington and Haiyan Zhang in the Metabolic Phenotyping Core at Virginia Tech for their technical assistance.
Funding: The study was funded by NIH R01MH105447-01 (SMC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
- Arias N, Mendez M, Vallejo G, Arias JL, 2015. Finding the place without the whole: Timeline involvement of brain regions. Brain Res. 1625, 18–28. 10.1016/j.brainres.2015.08.021 [DOI] [PubMed] [Google Scholar]
- Beckervordersandforth R, Ebert B, Schaffner I, Moss J, Fiebig C, Shin J, Moore DL, Ghosh L, Trinchero MF, Stockburger C, Friedland K, Steib K, von Wittgenstein J, Keiner S, Redecker C, Holter SM, Xiang W, Wurst W, Jagasia R, Schinder AF, Ming G-L, Toni N, Jessberger S, Song H, Lie DC, 2017. Role of Mitochondrial Metabolism in the Control of Early Lineage Progression and Aging Phenotypes in Adult Hippocampal Neurogenesis. Neuron 93, 560–573.e6. 10.1016/j.neuron.2016.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles RG, Burnett BB, Gleditsch K, Wong S, Guedalia A, Kaariainen A, Eloed J, Stern A, Brumm V, 2005. A high predisposition to depression and anxiety in mothers and other matrilineal relatives of children with presumed maternally inherited mitochondrial disorders. Am. J. Med. Genet. B. Neuropsychiatr. Genet 137B, 20–24. 10.1002/ajmg.b.30199 [DOI] [PubMed] [Google Scholar]
- Celikel FC, Kose S, Cumurcu BE, Erkorkmaz U, Sayar K, Borckardt JJ, Cloninger CR, 2009. Cloninger’s temperament and character dimensions of personality in patients with major depressive disorder. Compr. Psychiatry 50, 556–561. 10.1016/j.comppsych.2008.11.012 [DOI] [PubMed] [Google Scholar]
- Clinton S, Miller S, Watson SJ, Akil H, 2008. Prenatal stress does not alter innate novelty-seeking behavioral traits, but differentially affects individual differences in neuroendocrine stress responsivity. Psychoneuroendocrinology 33, 162–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton SM, Kerman IA, Orr HR, Bedrosian TA, Abraham AD, Simpson DN, Watson SJ, Akil H, 2011a. Pattern of forebrain activation in high novelty-seeking rats following aggressive encounter. Brain Res. 1422, 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton SM, Stead JD, Miller S, Watson SJ, Akil H, 2011b. Developmental underpinnings of differences in rodent novelty-seeking and emotional reactivity. Eur. J. Neurosci 34, 994–1005. 10.1111/j.1460-9568.2011.07811.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton SM, Watson SJ, Akil H, 2014. High novelty-seeking rats are resilient to negative physiological effects of the early life stress. Stress 17, 97–107. 10.3109/10253890.2013.850670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JL, Ata AE, Jackson NL, Rahn EJ, Ramaker RC, Cooper S, Kerman IA, Clinton SM, 2017. Differential stress induced c-Fos expression and identification of region-specific miRNA-mRNA networks in the dorsal raphe and amygdala of high-responder/low-responder rats. Behav. Brain Res. 319, 110–123. 10.1016/j.bbr.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JL, Glover ME, Pugh PC, Fant AD, Simmons RK, Akil H, Kerman IA, Clinton SM, 2015. Maternal Style Selectively Shapes Amygdalar Development and Social Behavior in Rats Genetically Prone to High Anxiety. Dev. Neurosci 10.1159/000374108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JA, Gowl BA, Westenbroek C, Clinton SM, Akil H, Becker JB, 2011. Effects of a selectively bred novelty-seeking phenotype on the motivation to take cocaine in male and female rats. Biol. Sex Differ. 2, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperfain AB, Zhang ZL, Kennedy JL, Goncalves VF, 2018. The Complex Interaction of Mitochondrial Genetics and Mitochondrial Pathways in Psychiatric Disease. Mol. neuropsychiatry 4, 52–69. 10.1159/000488031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BA, Clinton SM, Akil H, Becker JB, 2008. The effects of novelty-seeking phenotypes and sex differences on acquisition of cocaine self-administration in selectively bred High-Responder and Low-Responder rats. Pharmacol. Biochem. Behav 90, 331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vasconcellos APS, Zugno AI, Dos Santos AHDP, Nietto FB, Crema LM, Goncalves M, Franzon R, de Souza Wyse AT, da Rocha ER, Dalmaz C, 2005. Na+,K(+)-ATPase activity is reduced in hippocampus of rats submitted to an experimental model of depression: effect of chronic lithium treatment and possible involvement in learning deficits. Neurobiol. Learn. Mem 84, 102–110. 10.1016/j.nlm.2005.05.002 [DOI] [PubMed] [Google Scholar]
- Della FP, Abelaira HM, Reus GZ, Santos MAB dos Tomaz DB, Antunes AR, Scaini G, Morais MOS, Streck EL, Quevedo J, 2013. Treatment with tianeptine induces antidepressive-like effects and alters the neurotrophin levels, mitochondrial respiratory chain and cycle Krebs enzymes in the brain of maternally deprived adult rats. Metab. Brain Dis.28, 93–105. 10.1007/s11011-012-9375-x [DOI] [PubMed] [Google Scholar]
- Dragunow M, Faull R, 1989. The use of c-fos as a metabolic marker in neuronal pathway tracing. J. Neurosci. Methods 29, 261–265. [DOI] [PubMed] [Google Scholar]
- Eigsti IM, Zayas V, Mischel W,Shoda Y, Ayduk O, Dadlani MB, Davidson MC, Lawrence Aber J, Casey BJ, 2006. Predicting cognitive control from preschool to late adolescence and young adulthood. Psychol Sci 17, 478–484. [DOI] [PubMed] [Google Scholar]
- Filiou MD, Asara JM, Nussbaumer M, Teplytska L, Landgraf R, Turck CW, 2014. Behavioral extremes of trait anxiety in mice are characterized by distinct metabolic profiles. J. Psychiatr. Res 58, 115–122. 10.1016/j.jpsychires.2014.07.019 [DOI] [PubMed] [Google Scholar]
- Filiou MD, Zhang Y, Teplytska L, Reckow S, Gormanns P, Maccarrone G, Frank E, Kessler MS, Hambsch B, Nussbaumer M, Bunck M, Ludwig T, Yassouridis A, Holsboer F, Landgraf R, Turck CW, 2011. Proteomics and metabolomics analysis of a trait anxiety mouse model reveals divergent mitochondrial pathways. Biol. Psychiatry 70, 1074–1082. 10.1016/j.biopsych.2011.06.009 [DOI] [PubMed] [Google Scholar]
- Flagel SB, Waselus M, Clinton SM, Watson SJ, Akil H, 2014. Antecedents and consequences of drug abuse in rats selectively bred for high and low response to novelty. Neuropharmacology 76, 425–436. 10.1016/j.neuropharm.2013.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried NT, Moffat C, Seifert EL, Oshinsky ML, 2014. Functional mitochondrial analysis in acute brain sections from adult rats reveals mitochondrial dysfunction in a rat model of migraine. Am. J. Physiol. Cell Physiol. 307, C1017–30. 10.1152/ajpcel1.00332.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fuster MJ, Parsegian A, Watson SJ, Akil H, Flagel SB, 2017. Adolescent cocaine exposure enhances goal-tracking behavior and impairs hippocampal cell genesis selectively in adult bred low-responder rats. Psychopharmacology (Berl). 234, 1293–1305. 10.1007/S00213-017-4566-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fuster MJ, Perez JA, Clinton SM, Watson SJ, Akil H, 2010. Impact of cocaine on adult hippocampal neurogenesis in an animal model of differential propensity to drug abuse. Eur. J. Neurosci 31, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover ME, Clinton SM, 2016. Of rodents and humans: A comparative review of the neurobehavioral effects of early life SSRI exposure in preclinical and clinical research. Int. J. Dev. Neurosci 51, 50–72. 10.1016/j.ijdevneu.2016.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Lima F, Sadile AG, 2000. Network operations revealed by brain metabolic mapping in a genetic model of hyperactivity and attention deficit: the naples high- and low excitability rats. Neurosci. Biobehav. Rev 24, 157–160. [DOI] [PubMed] [Google Scholar]
- Harro J, Kanarik M, Kaart T, Matrov D, Koiv K, Mallo T, Del Rio J, Tordera RM, Ramirez MJ, 2014. Revealing the cerebral regions and networks mediating vulnerability to depression: oxidative metabolism mapping of rat brain. Behav. Brain Res. 267, 83–94. 10.1016/j.bbr.2014.03.019 [DOI] [PubMed] [Google Scholar]
- Harro J, Kanarik M, Matrov D, Panksepp J, 2011. Mapping patterns of depression-related brain regions with cytochrome oxidase histochemistry: relevance of animal affective systems to human disorders, with a focus on resilience to adverse events. Neurosci. Biobehav. Rev 35, 1876–1889. 10.1016/j.neubiorev.2011.02.016 [DOI] [PubMed] [Google Scholar]
- Hescham S, Temel Y, Casaca-Carreira J, Arslantas K, Yakkioui Y, Blokland A, Jahanshahi A, 2014. A neuroanatomical analysis of the effects of a memory impairing dose of scopolamine in the rat brain using cytochrome c oxidase as principle marker. J. Chem. Neuroanat 59–60, 1–7. 10.1016/j.jchemneu.2014.04.001 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA, 2009a. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA, 2009b. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Ishino H, Seno H, Ohguni S, Tanaka J, Kato Y, 1997. Psychiatric symptoms in a patient with diabetes mellitus associated with point mutation in mitochondrial DNA. Biol. Psychiatry 42, 1067–1069. [DOI] [PubMed] [Google Scholar]
- Kaczmarek L, 1995. Towards understanding of the role of transcription factors in learning processes. Acta Biochim. Pol. 42, 221–226. [PubMed] [Google Scholar]
- Kanarik M, Alttoa A, Matrov D, Koiv K, Sharp T, Panksepp J, Harro J, 2011. Brain responses to chronic social defeat stress: effects on regional oxidative metabolism as a function of a hedonic trait, and gene expression in susceptible and resilient rats. Eur. Neuropsychopharmacol 21, 92–107. 10.1016/j.euroneuro.2010.06.015 [DOI] [PubMed] [Google Scholar]
- Kanarik M, Harro J, 2018. Sociability trait and regional cerebral oxidative metabolism in rats: Predominantly nonlinear relations. Behav. Brain Res. 337, 186–192. 10.1016/j.bbr.2017.08.049 [DOI] [PubMed] [Google Scholar]
- Kanarik M, Matrov D, Koiv K, Eller M, Tonissaar M, Harro J, 2008. Changes in regional long-term oxidative metabolism induced by partial serotonergic denervation and chronic variable stress in rat brain. Neurochem. Int 52, 432–437. 10.1016/j.neuint.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Kerman IA, Clinton SM, Bedrosian TA, Abraham AD, Rosenthal DT, Akil H, Watson SJ, 2011. High novelty-seeking predicts aggression and gene expression differences within defined serotonergic cell groups. Brain Res. 1419, 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerman IA, Clinton SM, Simpson DN, Bedrosian TA, Bernard R, Akil H, Watson SJ, 2012. Inborn differences in environmental reactivity predict divergent diurnal behavioral, endocrine, and gene expression rhythms. Psychoneuroendocrinology 37, 256–269. 10.1016/j.psyneuen.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch M, Pilz G-A, Ghesquiere B, Kovacs WJ, Wegleiter T, Moore DL, Hruzova M, Zamboni N, Carmeliet P, Jessberger S, 2017. A Fatty Acid Oxidation-Dependent Metabolic Shift Regulates Adult Neural Stem Cell Activity. Cell Rep. 20, 2144–2155. 10.1016/j.celrep.2017.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarski JZ, McIntyre RS, Kennedy SH, Rafi-Tari S, Soczynska JK, Ketter TA, 2008. Volumetric neuroimaging investigations in mood disorders: bipolar disorder versus major depressive disorder. Bipolar Disord. 10, 1–37. 10.1111/j.1399-5618.2008.00435.x [DOI] [PubMed] [Google Scholar]
- Lee B-T, Seok J-H, Lee B-C, Cho SW, Yoon B-J, Lee K-U, Chae J-H, Choi l.-G., Ham B-J, 2008. Neural correlates of affective processing in response to sad and angry facial stimuli in patients with major depressive disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 32, 778–785. 10.1016/j.pnpbp.2007.12.009 [DOI] [PubMed] [Google Scholar]
- Madrigal JL, Olivenza R, Moro MA, Lizasoain I, Lorenzo P, Rodrigo J, Leza JC, 2001. Glutathione depletion, lipid peroxidation and mitochondrial dysfunction are induced by chronic stress in rat brain. Neuropsychopharmacology 24, 420–429. 10.1016/S0893-133X(00)00208-6 [DOI] [PubMed] [Google Scholar]
- Mallo T, Matrov D, Koiv K, Harro J, 2009. Effect of chronic stress on behavior and cerebral oxidative metabolism in rats with high or low positive affect. Neuroscience 164, 963–974. 10.1016/j.neuroscience.2009.08.041 [DOI] [PubMed] [Google Scholar]
- Marazziti D, Baroni S, Picchetti M, Landi P, Silvestri S, Vatteroni E, Catena Dell’Osso M, 2011. Mitochondrial alterations and neuropsychiatric disorders. Curr. Med. Chem 18, 4715–4721. [DOI] [PubMed] [Google Scholar]
- Matrov D, Kaart T, Lanfumey L, Maldonado R, Sharp T, Tordera RM, Kelly PA, Deakin B, Harro J, 2019. Cerebral oxidative metabolism mapping in four genetic mouse models of anxiety and mood disorders. Behav. Brain Res. 356, 435–443. 10.1016/j.bbr.2018.05.031 [DOI] [PubMed] [Google Scholar]
- Matrov D, Kolts I, Harro J, 2007. Cerebral oxidative metabolism in rats with high and low exploratory activity. Neurosci. Lett 413, 154–158. 10.1016/j.neulet.2006.11.076 [DOI] [PubMed] [Google Scholar]
- McCoy CR,Golf SR, Melendez-Ferro M, Perez-Costas E, Glover ME, Jackson NL, Stringfellow SA, Pugh PC, Fant AD, Clinton SM, 2016. Altered metabolic activity in the developing brain of rats predisposed to high versus low depression-like behavior. Neuroscience 324, 469–484. 10.1016/j.neuroscience.2016.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy CR, Jackson NL, Day J, Clinton SM, 2017. Genetic predisposition to high anxiety- and depression-like behavior coincides with diminished DNA methylation in the adult rat amygdala. Behav. Brain Res. 320, 165–178. 10.1016/j.bbr.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez-Ferro M, Rice MW, Roberts RC, Perez-Costas E, 2013. An accurate method for the quantification of cytochrome C oxidase in tissue sections. J Neurosci Methods 214, 156–162. 10.1016/j.jneumeth.2013.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Lopez M, Mendez M, Lopez L, Arias JL, 2009. Spatial working memory in Wistar rats: brain sex differences in metabolic activity. Brain Res. Bull. 79, 187–192. 10.1016/j.brainresbull.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Muir J, Lopez J, Bagot RC, 2019. Wiring the depressed brain: optogenetic and chemogenetic circuit interrogation in animal models of depression. Neuropsychopharmacology 44, 1013–1026. 10.1038/s41386-018-0291-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya JD, Pauly JR, Sullivan PG, 2009. The optimal dosage and window of opportunity to maintain mitochondrial homeostasis following traumatic brain injury using the uncoupler FCCP. Exp. Neurol 218, 381–389. 10.1016/j.expneurol.2009.05.023 [DOI] [PubMed] [Google Scholar]
- Perez JA, Clinton SM, Turner CA, Watson SJ, Akil H, 2009. A new role for FGF2 as an endogenous inhibitor of anxiety. J Neurosci 29, 6379–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, McManus MJ, Gray JD, Nasca C, Moffat C, Kopinski PK, Seifert EL, McEwen BS, Wallace DC, 2015. Mitochondrial functions modulate neuroendocrine, metabolic, inflammatory, and transcriptional responses to acute psychological stress. Proc. Natl. Acad. Sci. U. S. A 112, E6614–E6623. 10.1073/pnas.1515733112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reale D, Reader SM, Sol D, McDougall PT, Dingemanse NJ, 2007. Integrating animal temperament within ecology and evolution. Biol. Rev. Camb. Philos. Soc 82, 291–318. 10.1111/j.1469-185X.2007.00010.x [DOI] [PubMed] [Google Scholar]
- Rezin GT, Cardoso MR, Goncalves CL, Scaini G, Fraga DB, Riegel RE, Comim CM, Quevedo J, Streck EL, 2008. Inhibition of mitochondrial respiratory chain in brain of rats subjected to an experimental model of depression. Neurochem. Int 53, 395–400. 10.1016/j.neuint.2008.09.012 [DOI] [PubMed] [Google Scholar]
- Robertson HA, 1992. Immediate-early genes, neuronal plasticity, and memory. Biochem. Cell Biol. 70, 729–737. [DOI] [PubMed] [Google Scholar]
- Roosevelt TS, Ruhmann-Wennhold A, Nelson DH, 1973. Adrenal corticosteroid effects upon rat brain mitochondrial metabolism. Endocrinology 93, 619–625. 10.1210/endo-93-3-619 [DOI] [PubMed] [Google Scholar]
- Sauerbeck A, Pandya J, Singh I, Bittman K, Readnower R, Bing G, Sullivan P, 2011. Analysis of regional brain mitochondrial bioenergetics and susceptibility to mitochondrial inhibition utilizing a microplate based system. J. Neurosci. Methods 198, 36–43. 10.1016/j.jneumeth.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW, 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumake J, Edwards E, Gonzalez-Lima F, 2002. Dissociation of septo-hippocampal metabolism in the congenitally helpless rat. Neuroscience 114, 373–377. [DOI] [PubMed] [Google Scholar]
- Shumake J, Edwards E, Gonzalez-Lima F, 2001. Hypermetabolism of paraventricular hypothalamus in the congenitally helpless rat. Neurosci. Lett. 311, 45–48. [DOI] [PubMed] [Google Scholar]
- Shumake J, Gonzalez-Lima F, 2003. Brain systems underlying susceptibility to helplessness and depression. Behav Cogn Neurosci Rev 2, 198–221. [DOI] [PubMed] [Google Scholar]
- Simmons RK, Howard JL, Simpson DN, Akil H, Clinton SM, 2012. DNA methylation in the developing hippocampus and amygdala of anxiety-prone versus risk-taking rats. Dev. Neurosci 34, 58–67. 10.1159/000336641000336641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivey JM, Padilla E, Shumake JD, Gonzalez-Lima F, 2011. Effects of maternal separation, early handling, and gonadal sex on regional metabolic capacity of the preweanling rat brain. Brain Res. 1367, 198–206. 10.1016/j.brainres.2010.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead JD, Clinton S, Neal C, Schneider J, Jama A, Miller S, Vazquez DM, Watson SJ, Akil H, 2006. Selective breeding for divergence in novelty-seeking traits: heritability and enrichment in spontaneous anxiety-related behaviors. Behav Genet 36, 697–712. 10.1007/s10519-006-9058-7 [DOI] [PubMed] [Google Scholar]
- Turner CA, Clinton SM, Thompson RC, Watson SJ Jr., Akil H, 2011. Fibroblast growth factor-2 (FGF2) augmentation early in life alters hippocampal development and rescues the anxiety phenotype in vulnerable animals. Proc. Natl. Acad. Sci. U. S. A 108, 8021–8025. 10.1073/pnas.1103732108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videbech P, 2000. PET measurements of brain glucose metabolism and blood flow in major depressive disorder: a critical review. Acta Psychiatr. Scand. 101, 11–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.