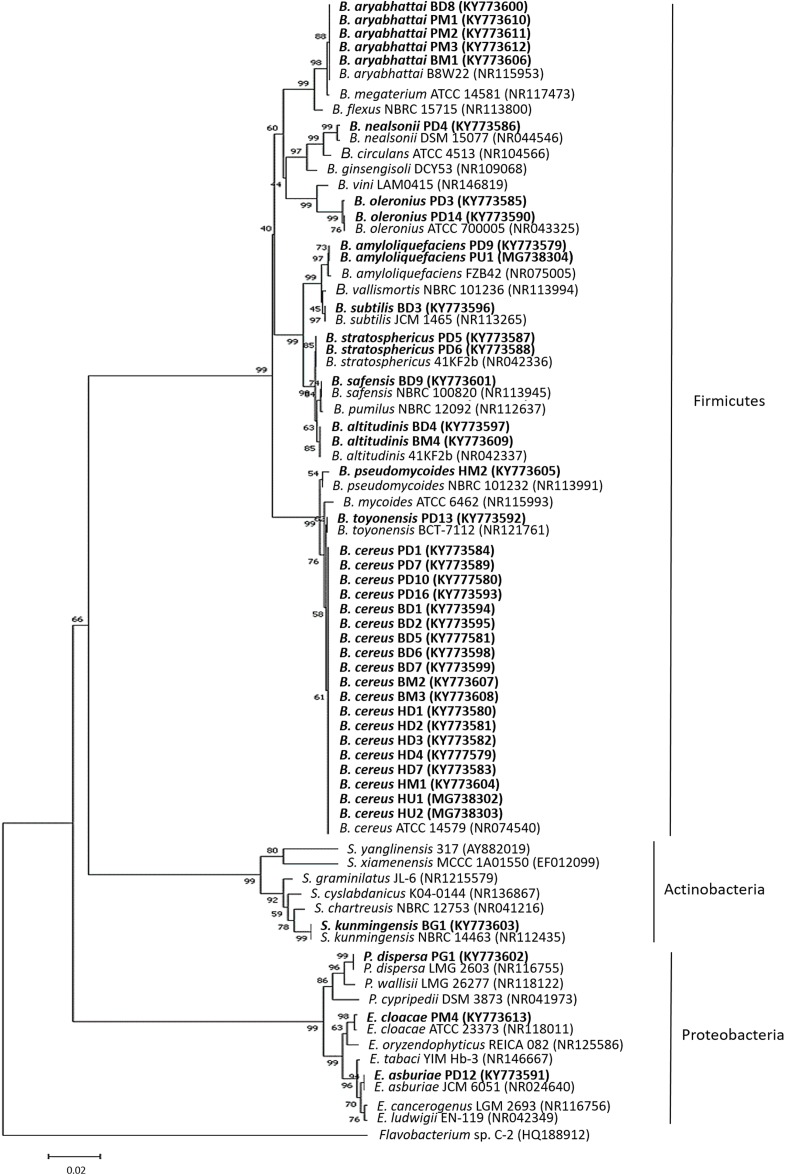
Figure 1. The 16S rRNA gene phylogenetic tree analysis of the bacterial isolates.
The evolutionary history was inferred using the neighbour-joining method. The isolates were aligned with closely related strains from GenBank. The accession numbers indicated within bracket. The optimal tree with the sum of branch length = 0.71705651 was shown. The analysis involved 76 nucleotide sequences. There were a total of 1,087 positions in the final dataset. Sequence from Flavobacterium sp. C-2 was used as an out group. The bolded text indicates bacteria isolated in this study. B, Bacillus; E, Enterobacter; P, Pantoea and S, Streptomyces.