Figure 3.
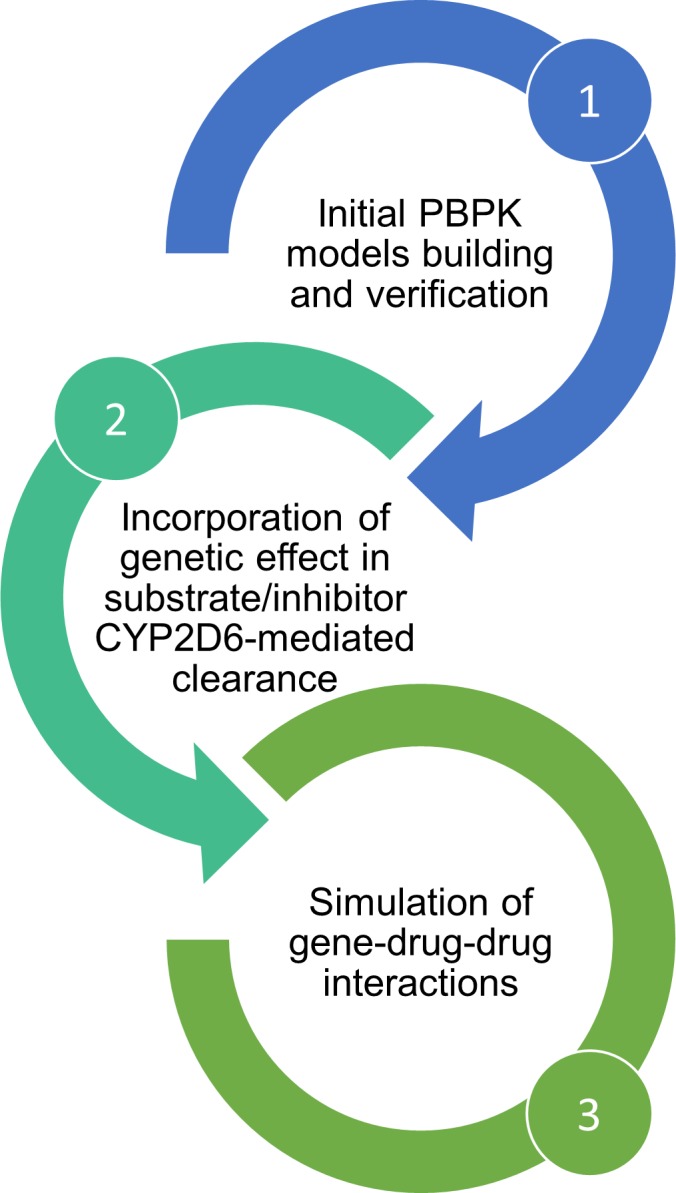
Study workflow. In a first step, physiologically‐based pharmacokinetic (PBPK) models for substrates and inhibitors were built, optimized, or used unchanged from verified library compounds in Simcyp version 17. In a second step, the Cytochrome P450 2D6 (CYP2D6) genotype‐dependent CYP2D6‐mediated clearance of substrate and inhibitors were estimated from existing in vivo data in humans. Then the third step consisted in the simulations of genotype‐dependent drug–drug interactions (DDIs) to compare with existing DDI trials.
