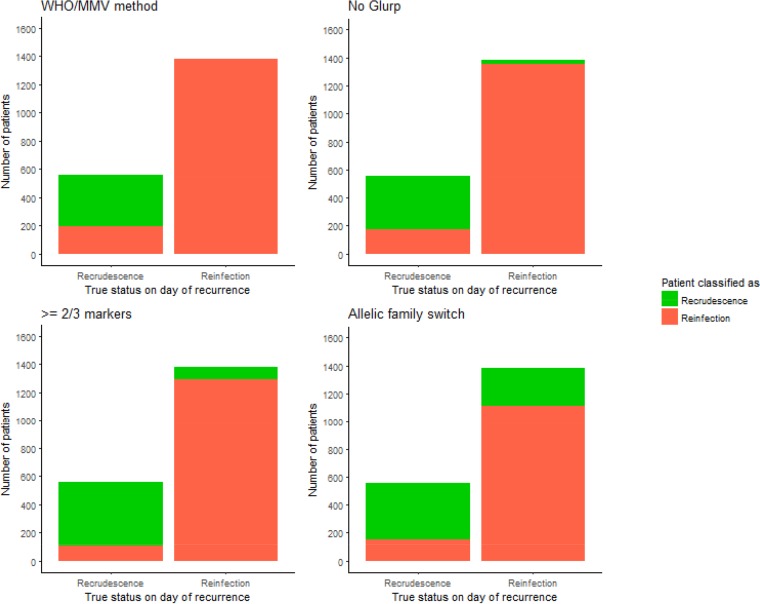
FIG 3.
Figure showing the ability of the various molecular correction algorithms to correctly classify patients with recurrent malaria. The data are for DHA-PPQ with a 42-day follow-up obtained with an FOI of 8 (i.e., used to obtain the results shown at FOI = 8 in Fig. 2). The multiplicity of infection (MOI) is drawn from data from Tanzania—a relatively high-transmission area. The x axis shows the true status of patients on the day of recurrence (i.e., reinfection or recrudescence), and the color coding shows how these patients were classified by each algorithm. The WHO/MMV-recommended algorithm correctly classifies nearly all reinfections, but misclassifies around one-third of recrudescences. The no-glurp algorithm is similar to the WHO/MMV one; it misclassifies only a small number of reinfections, but misclassifies around a third of recrudescences. The ≥2/3 markers algorithm had fewer misclassifications and was also more balanced (i.e., misclassified similar proportions of both reinfections and recrudescences). Finally, the allelic family switch algorithm correctly classifies a large proportion of recrudescences but misclassifies around half of reinfections.