Figure 4.
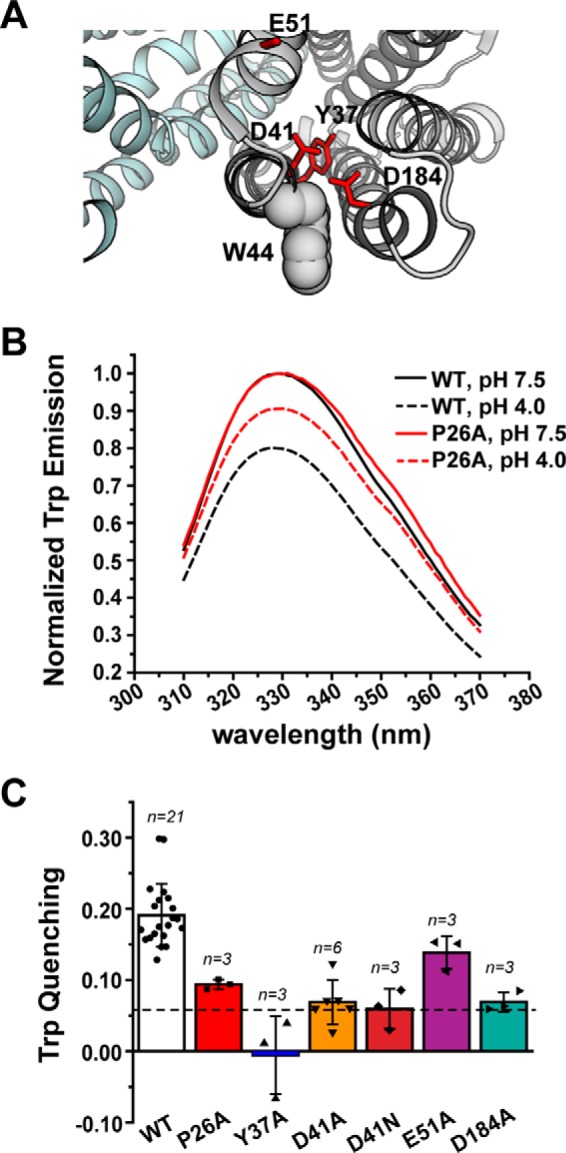
NTD amino acid substitutions disrupt the pattern of H+-dependent Trp quenching. A, structure of PfMATE (PDB code 3VVN), illustrating the location of Trp-44 (shown in space-filling representation) relative to the NTD variants. B, Trp fluorescence is quenched for WT PfMATE at pH 4 (black dashed trace), which is reduced by introduction of P26A (red dashed trace). The P26A spectra are shown as a representative dataset. Spectra were acquired at 23 °C in β-DDM buffer. C, profile of Trp quenching for the NTD variants. The bar plot highlights the mean ± S.D. for the indicated number of measurements. One-way ANOVA indicated that the population means were significantly different at the 0.05 level: F (6, 35) = 20.29, p = 4.53 × 10−10. The horizontal dashed line indicates the quenching observed for the W44C variant. The color code is the same as in Fig. 2.
